"mannosidase, alpha, class 2A, member 2"
ZFIN 
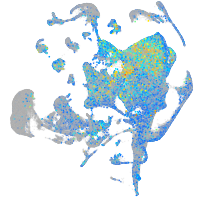
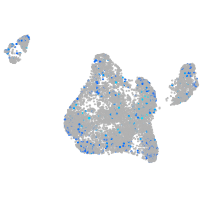
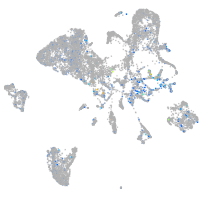
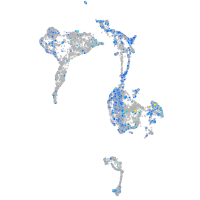
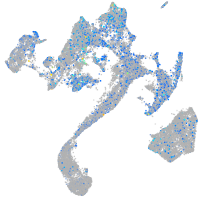
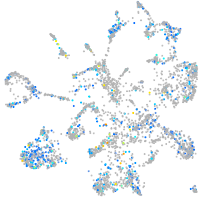
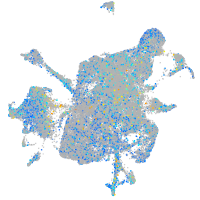
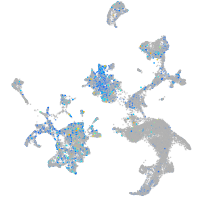
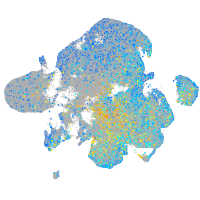
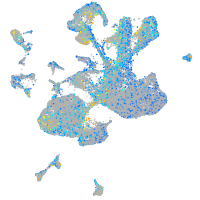
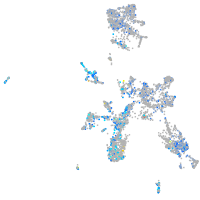
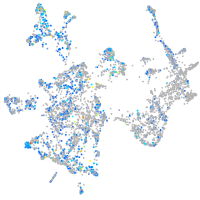
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pax6b | 0.154 | gapdh | -0.092 |
| XLOC-029934 | 0.153 | aldob | -0.080 |
| wdr17 | 0.140 | ahcy | -0.078 |
| SLCO3A1 (1 of many) | 0.139 | gamt | -0.078 |
| LO018020.2 | 0.139 | fabp2 | -0.076 |
| fam189a1 | 0.136 | gatm | -0.075 |
| scg3 | 0.136 | gpx4a | -0.071 |
| LOC110437922 | 0.136 | fbp1b | -0.070 |
| cga | 0.134 | apoc2 | -0.070 |
| neurod1 | 0.131 | gstp1 | -0.069 |
| tshr | 0.129 | apoa4b.1 | -0.068 |
| CABZ01066921.1 | 0.126 | eno3 | -0.064 |
| elfn2a | 0.125 | scp2a | -0.063 |
| NPFFR2 | 0.125 | afp4 | -0.062 |
| vat1 | 0.125 | gstt1a | -0.061 |
| mir7a-1 | 0.124 | cox8b | -0.061 |
| LOC100537384 | 0.123 | eef1da | -0.061 |
| insm1a | 0.122 | gcshb | -0.061 |
| syt1a | 0.122 | rbp2a | -0.060 |
| myt1b | 0.118 | cox7a1 | -0.059 |
| BX005462.2 | 0.118 | krt92 | -0.059 |
| si:dkey-27j5.5 | 0.118 | pla2g12b | -0.059 |
| myt1a | 0.118 | cdo1 | -0.059 |
| cacng4b | 0.117 | gsta.1 | -0.059 |
| si:ch211-254n4.3 | 0.117 | apoa1b | -0.058 |
| kcnt2 | 0.116 | dap | -0.058 |
| atpv0e2 | 0.116 | apobb.1 | -0.057 |
| isl1 | 0.116 | glud1b | -0.056 |
| rnasekb | 0.116 | suclg1 | -0.056 |
| pcsk1nl | 0.115 | sult2st2 | -0.056 |
| jpt1b | 0.115 | ugt1a7 | -0.056 |
| si:ch1073-429i10.3.1 | 0.114 | rmdn1 | -0.056 |
| scgn | 0.114 | mat1a | -0.056 |
| apc2 | 0.113 | zgc:163080 | -0.055 |
| CR392036.1 | 0.113 | cx32.3 | -0.054 |