"leucine zipper, putative tumor suppressor 2a"
ZFIN 
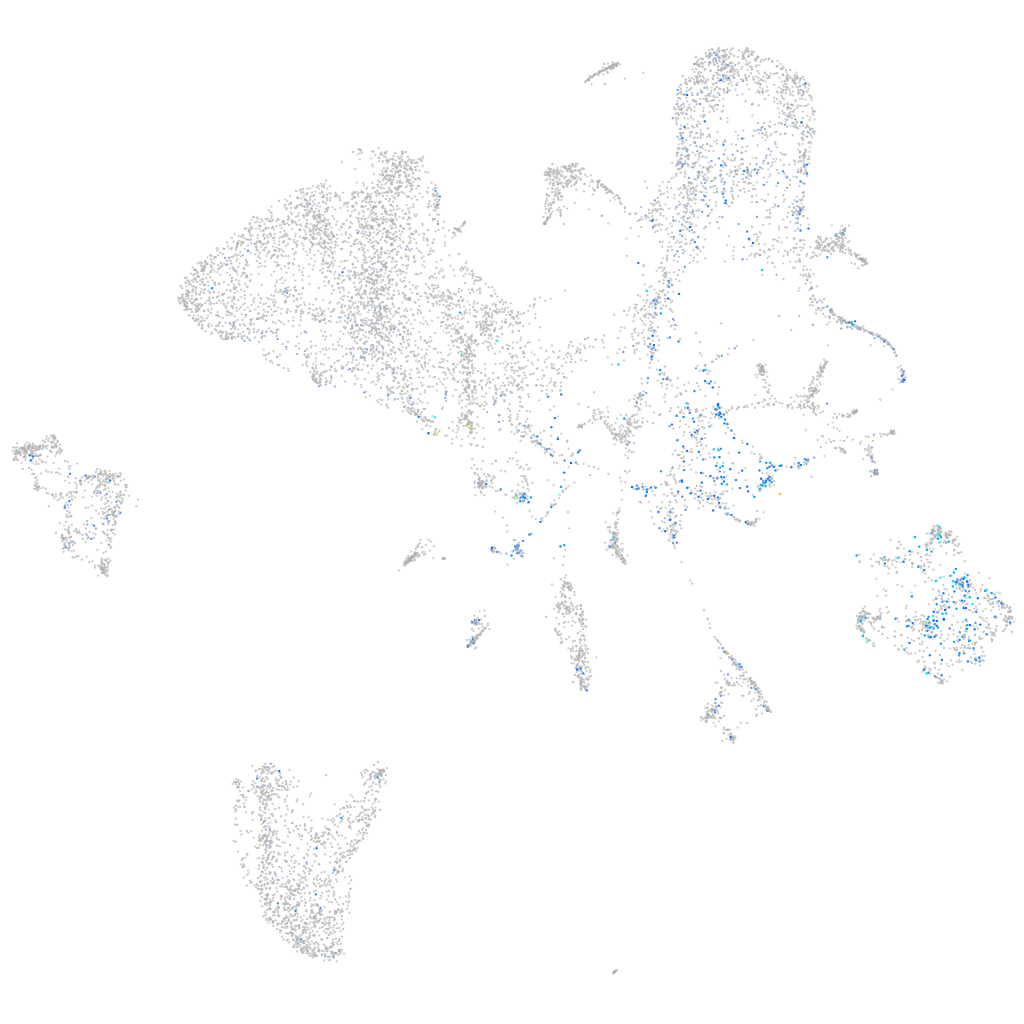
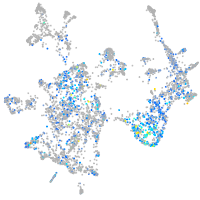
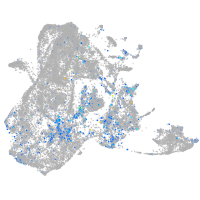
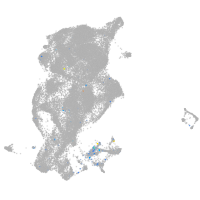
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdh6 | 0.265 | gamt | -0.173 |
| cx43.4 | 0.252 | bhmt | -0.169 |
| qkia | 0.249 | gapdh | -0.166 |
| nucks1a | 0.249 | ahcy | -0.164 |
| syncrip | 0.244 | gatm | -0.161 |
| hmga1a | 0.244 | agxtb | -0.154 |
| seta | 0.242 | mat1a | -0.153 |
| si:ch211-222l21.1 | 0.241 | apoa2 | -0.150 |
| marcksb | 0.241 | apoa1b | -0.150 |
| cbx5 | 0.240 | sod1 | -0.148 |
| khdrbs1a | 0.239 | pnp4b | -0.144 |
| ilf3b | 0.235 | rbp4 | -0.143 |
| cdx4 | 0.235 | grhprb | -0.140 |
| akap12b | 0.233 | gpx4a | -0.140 |
| hp1bp3 | 0.232 | zgc:123103 | -0.139 |
| acin1a | 0.230 | rbp2b | -0.139 |
| hspb1 | 0.230 | apom | -0.138 |
| si:ch73-281n10.2 | 0.230 | fabp10a | -0.137 |
| znfl2a | 0.230 | serpina1 | -0.136 |
| hnrnpaba | 0.229 | dap | -0.136 |
| cirbpa | 0.228 | nupr1b | -0.136 |
| chaf1a | 0.228 | aqp12 | -0.136 |
| smc1al | 0.227 | ces2 | -0.135 |
| snrpd3l | 0.227 | serpina1l | -0.135 |
| cirbpb | 0.227 | fbp1b | -0.134 |
| si:ch73-1a9.3 | 0.226 | pgm1 | -0.133 |
| ncl | 0.226 | kng1 | -0.133 |
| snrpb | 0.225 | ttc36 | -0.133 |
| hapln1b | 0.225 | hao1 | -0.133 |
| h3f3d | 0.224 | fgb | -0.132 |
| smarca4a | 0.224 | tfa | -0.132 |
| cbx1a | 0.222 | fgg | -0.132 |
| tgif1 | 0.222 | ambp | -0.132 |
| cbx3a | 0.222 | gnmt | -0.131 |
| id3 | 0.222 | LOC110437731 | -0.131 |