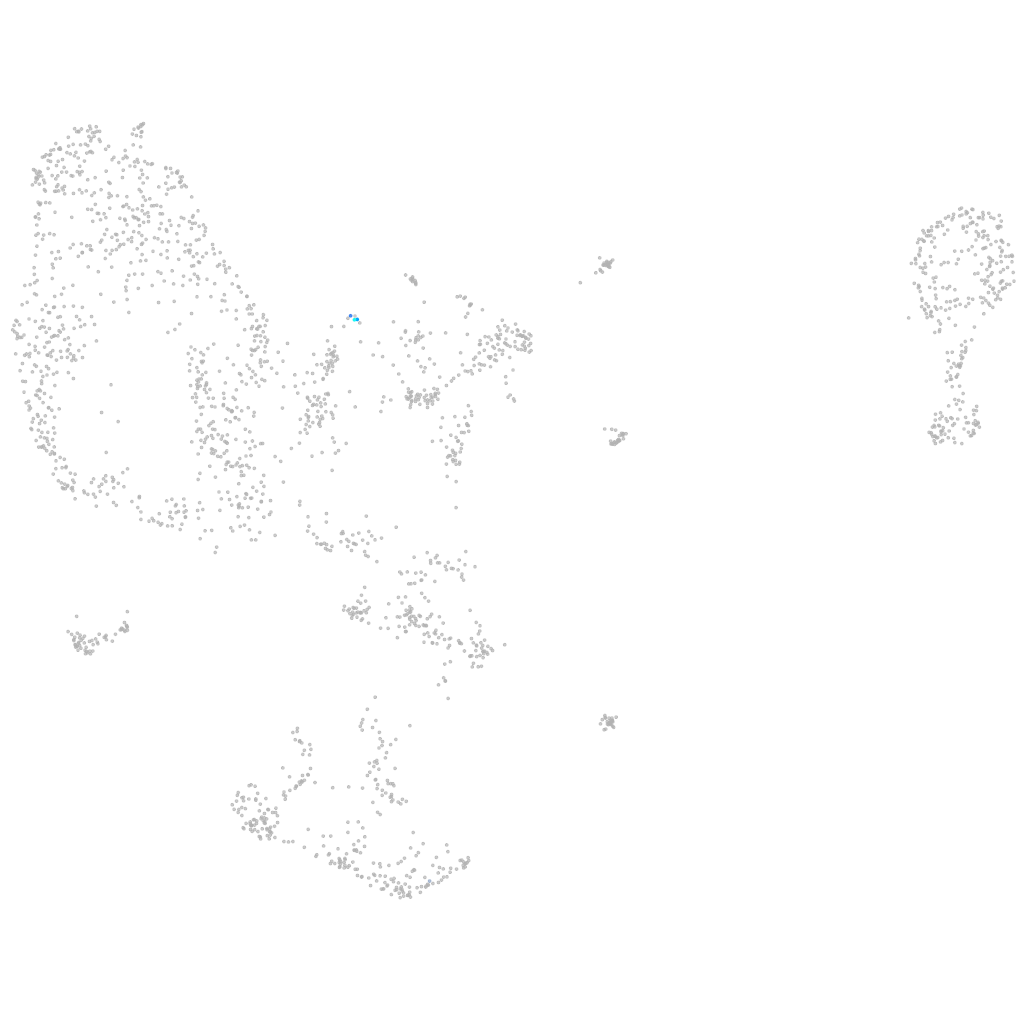
lymphatic vessel endothelial hyaluronic receptor 1b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sh3bp2 | 0.960 | snrpe | -0.035 |
| si:ch211-33e4.2 | 0.824 | NC-002333.4 | -0.035 |
| LOC108182616 | 0.809 | rps12 | -0.034 |
| si:dkey-151m22.8 | 0.802 | top1l | -0.031 |
| CU463284.2 | 0.802 | snrpf | -0.029 |
| LOC101886371 | 0.802 | si:ch211-193l2.10 | -0.028 |
| LOC101886679 | 0.802 | ywhae1 | -0.028 |
| LOC103909948 | 0.802 | hmga1a | -0.028 |
| adra1d | 0.797 | hnrnpaba | -0.027 |
| itga8 | 0.741 | si:ch211-222l21.1 | -0.027 |
| si:ch211-14k19.8 | 0.738 | setb | -0.027 |
| bmp16 | 0.705 | rps11 | -0.027 |
| plvapb | 0.694 | fkbp3 | -0.027 |
| rca2.2 | 0.674 | hnrnpa1b | -0.026 |
| ecscr | 0.662 | apex1 | -0.026 |
| ptprb | 0.650 | psmb3 | -0.026 |
| mrc1a | 0.639 | abracl | -0.026 |
| exoc3l2a | 0.627 | snrpd1 | -0.025 |
| gpr182 | 0.622 | si:ch73-281n10.2 | -0.025 |
| kdrl | 0.621 | psma6a | -0.025 |
| cd81b | 0.606 | ndrg3a | -0.025 |
| calcrlb | 0.585 | cldnc | -0.025 |
| agtr1b | 0.582 | ptmab | -0.025 |
| pecam1 | 0.575 | si:ch73-1a9.3 | -0.025 |
| thsd1 | 0.563 | sf3b2 | -0.025 |
| zgc:172122 | 0.552 | rps18 | -0.025 |
| fam167b | 0.550 | rpl22l1 | -0.025 |
| slc6a4a | 0.538 | rbx1 | -0.025 |
| limch1b | 0.517 | sub1a | -0.025 |
| CABZ01067979.1 | 0.506 | cstf2 | -0.024 |
| loxl1 | 0.505 | ifrd2 | -0.024 |
| kdr | 0.498 | emc6 | -0.024 |
| si:ch211-113d11.5 | 0.496 | bcas2 | -0.024 |
| tek | 0.496 | tomm40l | -0.024 |
| myct1a | 0.496 | taf10 | -0.024 |