"LysM, putative peptidoglycan-binding, domain containing 2"
ZFIN 
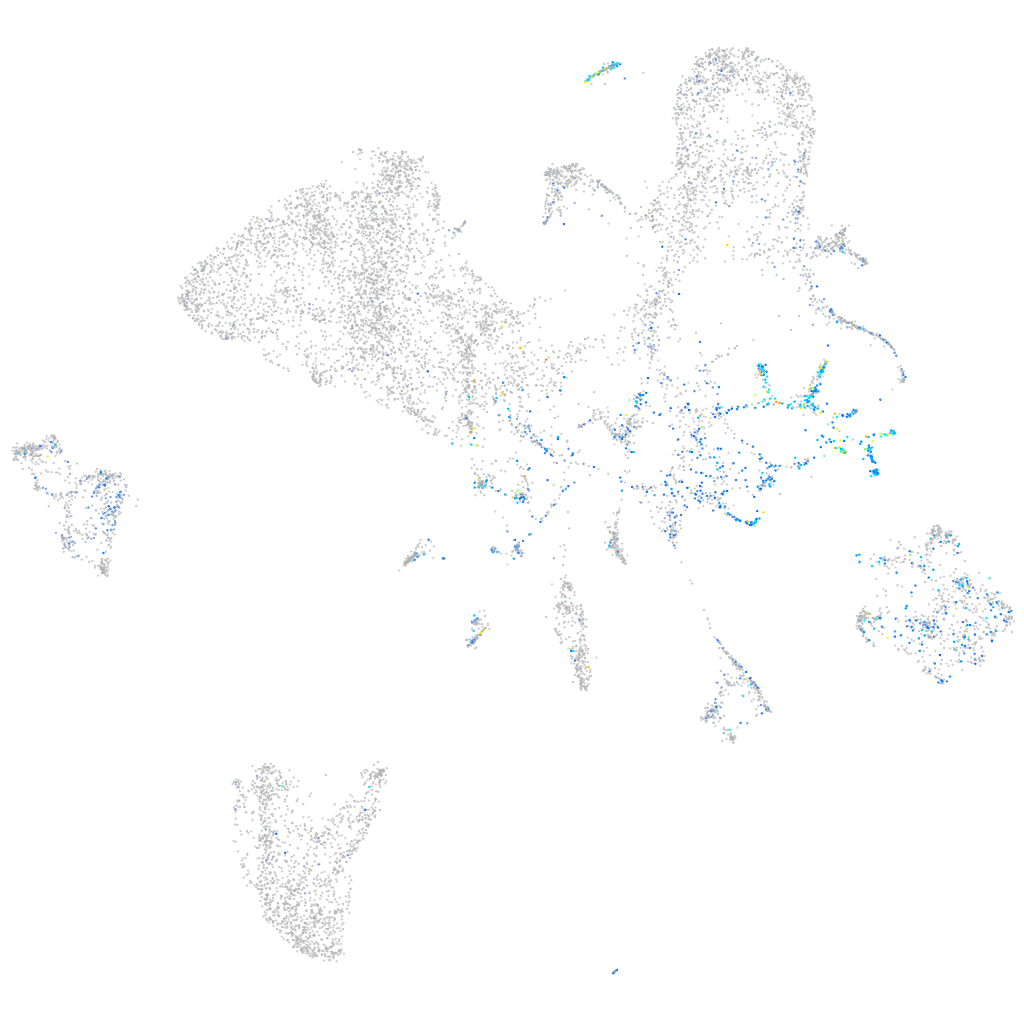
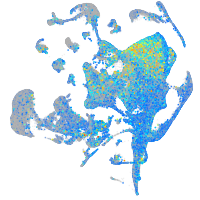
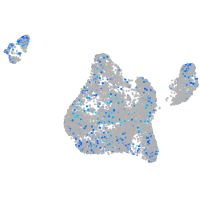
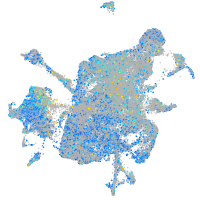
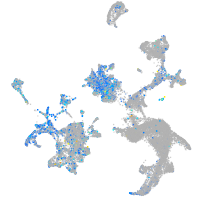
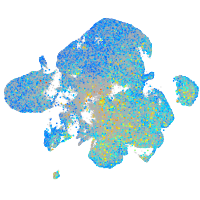
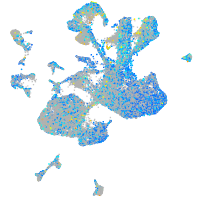
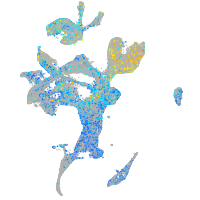
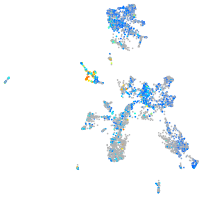
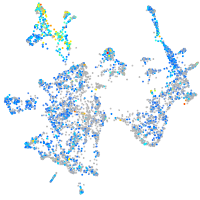
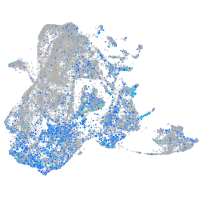
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.521 | gapdh | -0.302 |
| neurod1 | 0.476 | ahcy | -0.289 |
| insm1a | 0.432 | gamt | -0.286 |
| pax6b | 0.432 | gatm | -0.263 |
| pcsk1nl | 0.410 | aldob | -0.255 |
| insm1b | 0.398 | fbp1b | -0.249 |
| mir7a-1 | 0.392 | mat1a | -0.241 |
| scgn | 0.380 | nupr1b | -0.237 |
| egr4 | 0.376 | apoa4b.1 | -0.236 |
| id4 | 0.368 | bhmt | -0.235 |
| nkx2.2a | 0.365 | apoa1b | -0.233 |
| si:dkey-153k10.9 | 0.364 | cx32.3 | -0.227 |
| zgc:101731 | 0.364 | eno3 | -0.227 |
| mir375-2 | 0.359 | apoc2 | -0.227 |
| isl1 | 0.356 | scp2a | -0.223 |
| scg2a | 0.354 | gstt1a | -0.222 |
| jpt1b | 0.352 | glud1b | -0.222 |
| rprmb | 0.347 | aldh6a1 | -0.221 |
| vat1 | 0.346 | agxtb | -0.219 |
| pcsk1 | 0.345 | afp4 | -0.217 |
| myt1b | 0.344 | aldh7a1 | -0.216 |
| vamp2 | 0.342 | suclg1 | -0.210 |
| c2cd4a | 0.340 | gnmt | -0.207 |
| syt1a | 0.337 | gpx4a | -0.207 |
| kcnj11 | 0.335 | apoa2 | -0.204 |
| tox | 0.335 | suclg2 | -0.204 |
| fev | 0.331 | pnp4b | -0.203 |
| pygma | 0.326 | agxta | -0.202 |
| gpc1a | 0.322 | hdlbpa | -0.200 |
| rnasekb | 0.320 | aqp12 | -0.200 |
| tspan7 | 0.318 | dap | -0.199 |
| hmgn6 | 0.316 | apobb.1 | -0.198 |
| tspan7b | 0.315 | nipsnap3a | -0.198 |
| jagn1a | 0.310 | mgst1.2 | -0.197 |
| hmgb3a | 0.307 | tdo2a | -0.196 |