leucine rich repeat containing 39
ZFIN 
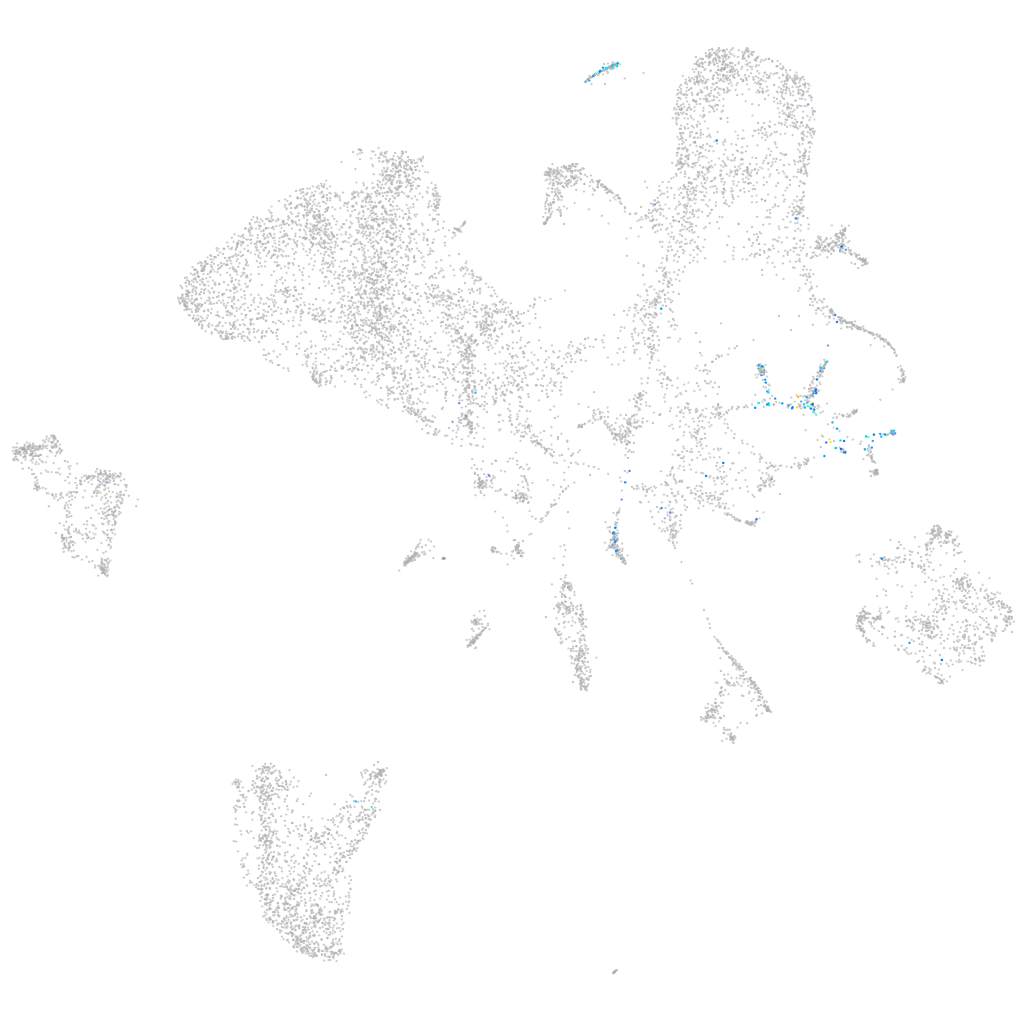
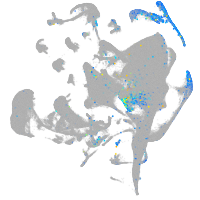
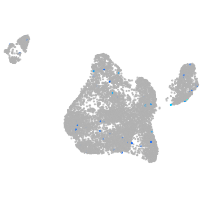
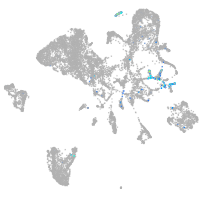
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.405 | ahcy | -0.124 |
| mir7a-1 | 0.373 | gamt | -0.110 |
| c2cd4a | 0.355 | fabp3 | -0.105 |
| neurod1 | 0.336 | gapdh | -0.101 |
| ghrl | 0.316 | aldob | -0.098 |
| pcsk1nl | 0.307 | fbp1b | -0.094 |
| mir375-2 | 0.303 | aldh6a1 | -0.094 |
| insm1a | 0.294 | nupr1b | -0.092 |
| sprn2 | 0.293 | gatm | -0.092 |
| pax6b | 0.292 | gstr | -0.092 |
| egr4 | 0.292 | mat1a | -0.091 |
| nkx2.2a | 0.291 | eif4ebp1 | -0.090 |
| pygma | 0.287 | si:dkey-16p21.8 | -0.090 |
| sst2 | 0.282 | bhmt | -0.088 |
| capsla | 0.281 | apoa4b.1 | -0.088 |
| insm1b | 0.279 | apoa1b | -0.086 |
| zgc:101731 | 0.275 | cx32.3 | -0.086 |
| pax4 | 0.274 | aldh7a1 | -0.085 |
| zgc:165481 | 0.269 | afp4 | -0.084 |
| syt1a | 0.269 | gstt1a | -0.083 |
| fev | 0.267 | apoc2 | -0.083 |
| pdyn | 0.267 | apoc1 | -0.083 |
| cacna1c | 0.258 | aqp12 | -0.082 |
| arxa | 0.255 | hdlbpa | -0.082 |
| pcsk1 | 0.252 | eef1da | -0.081 |
| lysmd2 | 0.249 | rps20 | -0.080 |
| dll4 | 0.249 | agxta | -0.080 |
| vat1 | 0.246 | mgst1.2 | -0.080 |
| hepacam2 | 0.245 | acadm | -0.080 |
| slc7a14a | 0.243 | eno3 | -0.080 |
| scgn | 0.241 | agxtb | -0.079 |
| si:dkey-153k10.9 | 0.241 | apoa2 | -0.079 |
| kcnj11 | 0.241 | scp2a | -0.079 |
| tmem63c | 0.241 | tfa | -0.079 |
| id4 | 0.240 | wu:fj16a03 | -0.078 |