leucine rich repeat containing 31
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural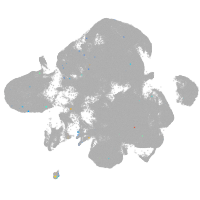 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm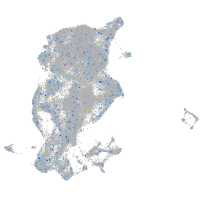 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| NAT16 (1 of many) | 0.300 | bhmt | -0.126 |
| dab2 | 0.288 | gatm | -0.123 |
| amn | 0.287 | aqp12 | -0.119 |
| si:dkey-28n18.9 | 0.283 | gamt | -0.112 |
| lrp2b | 0.282 | apoc1 | -0.105 |
| LOC100537272 | 0.282 | mat1a | -0.100 |
| fabp6 | 0.274 | cdo1 | -0.094 |
| cdaa | 0.272 | prss1 | -0.089 |
| ctsl.1 | 0.269 | pnp4b | -0.089 |
| cubn | 0.268 | cpb1 | -0.088 |
| tspan34 | 0.260 | ppdpfa | -0.088 |
| tfeb | 0.259 | apobb.1 | -0.088 |
| xirp1 | 0.257 | ela2l | -0.088 |
| sptbn5 | 0.256 | ctrl | -0.088 |
| si:ch211-139a5.9 | 0.255 | fep15 | -0.087 |
| ctsbb | 0.255 | si:dkey-56m19.5 | -0.087 |
| slc10a2 | 0.253 | cpa5 | -0.087 |
| snx2 | 0.251 | CELA1 (1 of many) | -0.087 |
| naga | 0.246 | rtn1a | -0.087 |
| si:ch211-202h22.9 | 0.243 | cel.1 | -0.087 |
| rab32b | 0.238 | ela2 | -0.086 |
| grn1 | 0.238 | sycn.2 | -0.086 |
| tmigd1 | 0.238 | prss59.1 | -0.085 |
| b3gnt3.4 | 0.238 | cpa4 | -0.085 |
| si:dkey-87o1.2 | 0.235 | cel.2 | -0.085 |
| krt222 | 0.232 | zgc:112160 | -0.085 |
| CABZ01112049.1 | 0.230 | ctrb1 | -0.084 |
| agr2 | 0.229 | si:ch211-240l19.5 | -0.084 |
| pllp | 0.225 | zgc:92744 | -0.084 |
| si:ch1073-443f11.2 | 0.225 | agxtb | -0.084 |
| slc15a2 | 0.224 | apoc2 | -0.083 |
| si:dkey-194e6.1 | 0.224 | zgc:136461 | -0.082 |
| rbm47 | 0.222 | ela3l | -0.082 |
| ddit3 | 0.220 | apoda.2 | -0.082 |
| zgc:153284 | 0.220 | c6ast4 | -0.082 |




