low density lipoprotein receptor-related protein associated protein 1
ZFIN 
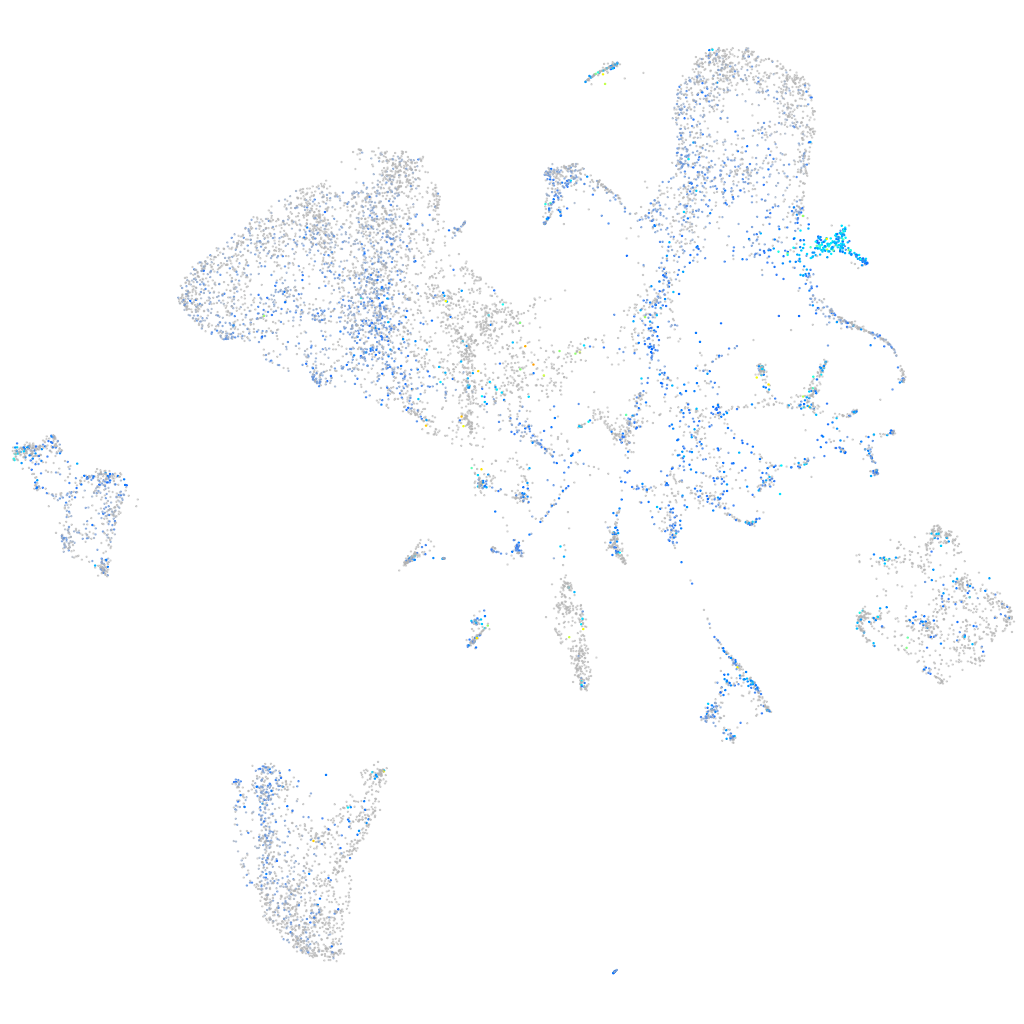
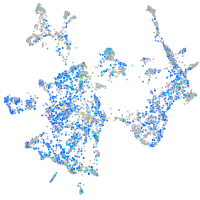
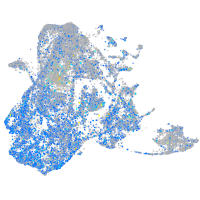
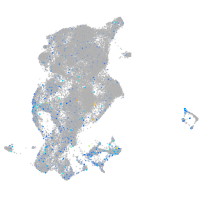
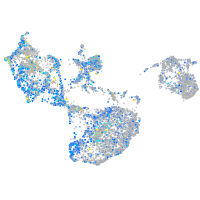
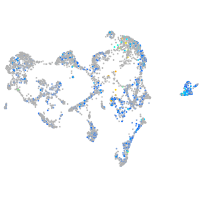
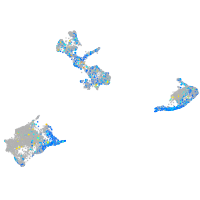
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dab2 | 0.372 | gatm | -0.178 |
| LOC100537272 | 0.369 | gamt | -0.174 |
| lrp2b | 0.364 | pnp4b | -0.172 |
| ctsbb | 0.361 | bhmt | -0.168 |
| amn | 0.352 | apoc1 | -0.156 |
| cubn | 0.348 | ucp1 | -0.153 |
| tfeb | 0.345 | mat1a | -0.148 |
| xirp1 | 0.331 | agxtb | -0.147 |
| sptbn5 | 0.323 | apobb.1 | -0.141 |
| naga | 0.319 | apoa1b | -0.139 |
| scpep1 | 0.310 | fetub | -0.137 |
| snx2 | 0.308 | cyp2n13 | -0.136 |
| cdaa | 0.308 | apoa2 | -0.134 |
| si:dkey-28n18.9 | 0.307 | cyp2ad2 | -0.130 |
| NAT16 (1 of many) | 0.301 | rbp4 | -0.128 |
| ctsl.1 | 0.296 | afp4 | -0.126 |
| si:dkey-194e6.1 | 0.293 | fabp10a | -0.125 |
| si:ch211-122f10.4 | 0.279 | ces2 | -0.124 |
| tpte | 0.278 | serpina1 | -0.122 |
| rab32b | 0.277 | aqp12 | -0.121 |
| si:ch211-139a5.9 | 0.270 | grhprb | -0.121 |
| slc15a2 | 0.269 | apom | -0.121 |
| fabp6 | 0.265 | faxdc2 | -0.120 |
| lgmn | 0.264 | comtd1 | -0.119 |
| CU499336.2 | 0.262 | apoc2 | -0.119 |
| dpep1 | 0.257 | dgat2 | -0.119 |
| tspan34 | 0.257 | cdo1 | -0.118 |
| tmigd1 | 0.254 | gapdh | -0.117 |
| slc6a18 | 0.254 | rbp2b | -0.117 |
| snx8a | 0.245 | LOC110437731 | -0.116 |
| slc10a2 | 0.241 | ttr | -0.115 |
| prss16 | 0.239 | zgc:123103 | -0.115 |
| grn1 | 0.239 | zgc:77439 | -0.114 |
| satb2 | 0.233 | rbp2a | -0.112 |
| cpvl | 0.231 | hpda | -0.112 |