low density lipoprotein receptor-related protein 1Aa
ZFIN 
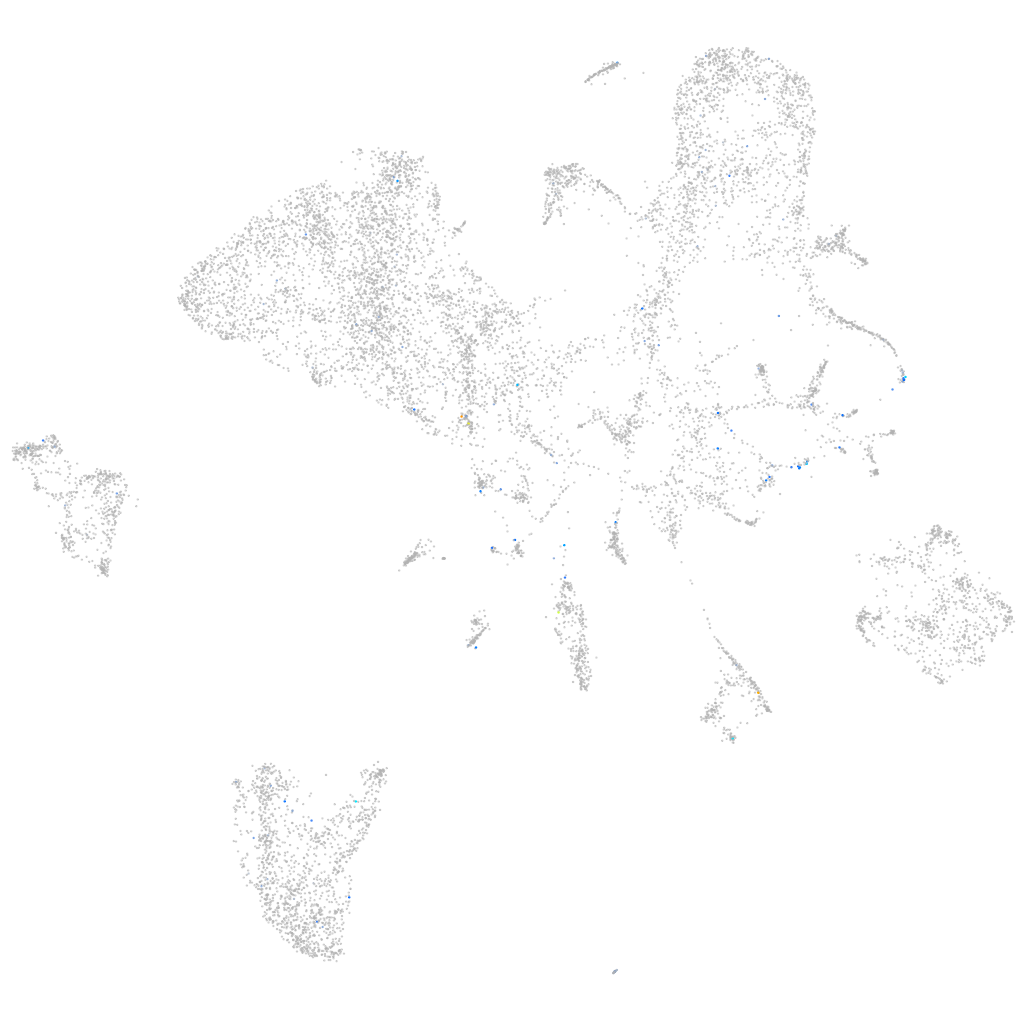
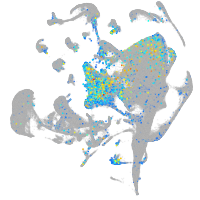
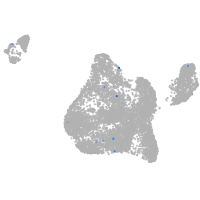
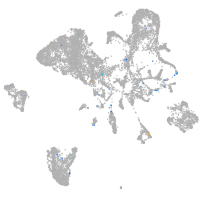
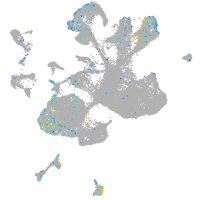
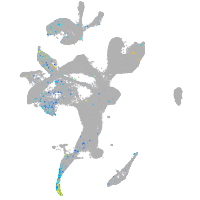
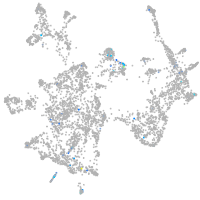
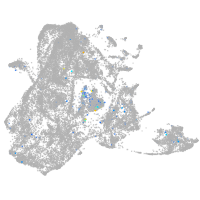
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-47f2.1 | 0.331 | eno3 | -0.047 |
| gpr17 | 0.321 | prdx2 | -0.047 |
| cntnap1 | 0.310 | gamt | -0.042 |
| slitrk3a | 0.277 | scp2a | -0.042 |
| vat1l | 0.214 | suclg1 | -0.042 |
| si:ch73-335m24.2 | 0.211 | cox7b | -0.042 |
| grm1a | 0.206 | COX5B | -0.041 |
| ntn2 | 0.199 | ahcy | -0.041 |
| thbs2b | 0.186 | acaa2 | -0.041 |
| LOC103911713 | 0.184 | aldob | -0.041 |
| fgf10a | 0.175 | zgc:193541 | -0.041 |
| unm-hu7912 | 0.174 | rmdn1 | -0.041 |
| CU657976.1 | 0.174 | mdh1aa | -0.040 |
| si:ch211-281p14.2 | 0.171 | gpx4a | -0.040 |
| admb | 0.170 | pklr | -0.040 |
| BX936284.1 | 0.167 | gstt1a | -0.040 |
| aldoca | 0.164 | abat | -0.040 |
| baiap3 | 0.161 | gapdh | -0.039 |
| adamtsl3 | 0.161 | hspe1 | -0.039 |
| dnah11 | 0.160 | sdr16c5b | -0.038 |
| FQ323119.1 | 0.159 | tdo2a | -0.038 |
| tspan5a | 0.158 | sod1 | -0.038 |
| tnr | 0.153 | shmt1 | -0.038 |
| lingo1a | 0.152 | cx32.3 | -0.038 |
| itgbl1 | 0.152 | fbp1b | -0.037 |
| si:ch73-7i4.2 | 0.152 | nipsnap3a | -0.037 |
| ttc12 | 0.151 | rdh1 | -0.036 |
| c7b | 0.149 | ugt1a7 | -0.036 |
| tmeff2a | 0.144 | sod2 | -0.036 |
| cldn19 | 0.144 | pgm1 | -0.036 |
| neto2a | 0.144 | uqcrfs1 | -0.036 |
| loxl1 | 0.143 | haao | -0.035 |
| FP016056.1 | 0.142 | eci2 | -0.035 |
| col8a2 | 0.139 | srd5a2a | -0.035 |
| LOC103908970 | 0.139 | g6pca.2 | -0.035 |