"LIM homeobox transcription factor 1, beta b"
ZFIN 
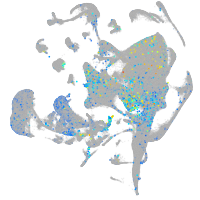
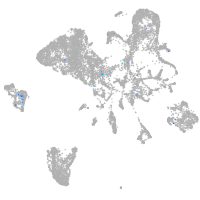
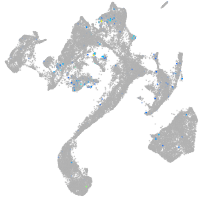
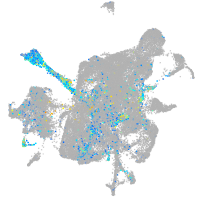
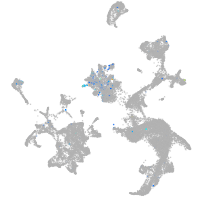
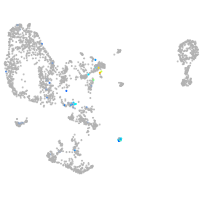
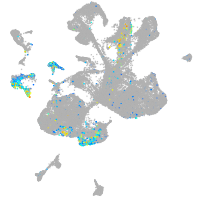
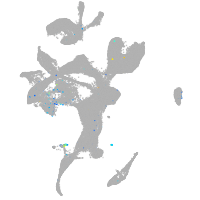
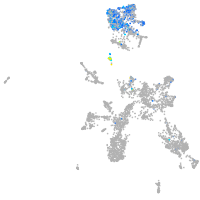
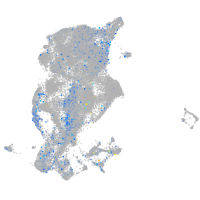
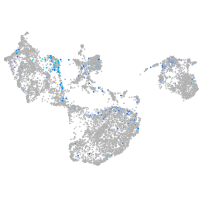
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR759836.1 | 0.316 | gapdh | -0.056 |
| LOC108179280 | 0.315 | gamt | -0.052 |
| si:ch73-205h11.1 | 0.299 | nupr1b | -0.048 |
| il2rb | 0.291 | aqp12 | -0.048 |
| grp | 0.246 | dap | -0.048 |
| pvalb8 | 0.238 | gatm | -0.048 |
| LOC101883110 | 0.237 | glud1b | -0.045 |
| nphs1 | 0.237 | scp2a | -0.044 |
| CR855375.1 | 0.230 | fbp1b | -0.044 |
| nphs2 | 0.227 | pcbd1 | -0.043 |
| ppp4r4 | 0.226 | kng1 | -0.042 |
| ano2 | 0.220 | upb1 | -0.042 |
| LOC100537721 | 0.218 | mat1a | -0.041 |
| tmtops2b | 0.218 | agxta | -0.041 |
| si:ch73-380l3.4 | 0.213 | grhprb | -0.041 |
| CABZ01073083.1 | 0.211 | pnp4b | -0.041 |
| otofa | 0.210 | cox7a1 | -0.040 |
| BX248128.2 | 0.209 | apoa4b.1 | -0.040 |
| gfral | 0.198 | cx32.3 | -0.040 |
| si:ch211-193i15.1 | 0.195 | cat | -0.040 |
| sgms2a | 0.191 | aldh1l1 | -0.040 |
| tph1a | 0.190 | suclg1 | -0.040 |
| dmrta2 | 0.188 | tdo2a | -0.040 |
| LOC100536671 | 0.185 | mst1 | -0.040 |
| C12orf75 | 0.181 | nipsnap3a | -0.039 |
| XLOC-011873 | 0.178 | abat | -0.039 |
| mkks | 0.178 | pklr | -0.039 |
| guca1c | 0.177 | mgst1.2 | -0.039 |
| XLOC-035750 | 0.170 | pgm1 | -0.039 |
| znf1043 | 0.170 | msrb2 | -0.039 |
| kcnab1a | 0.167 | hpda | -0.039 |
| TCERG1L | 0.165 | ddt | -0.038 |
| LOC556170 | 0.164 | zgc:92744 | -0.038 |
| wnt1 | 0.158 | sult2st2 | -0.038 |
| trim35-21 | 0.157 | ftcd | -0.038 |