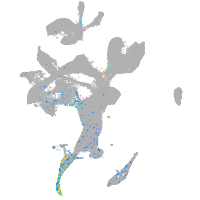
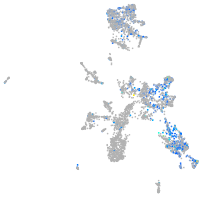
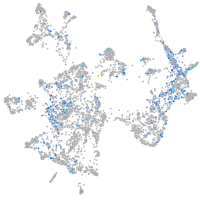
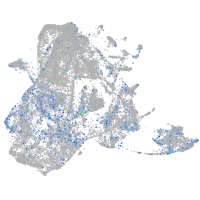
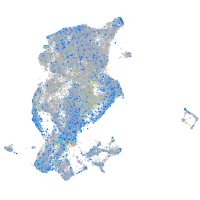
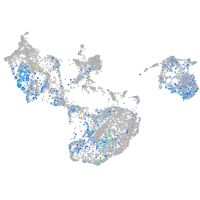
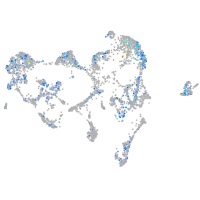
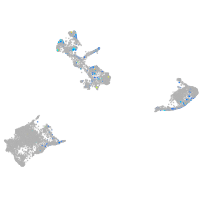
"lipase, endothelial"
ZFIN 
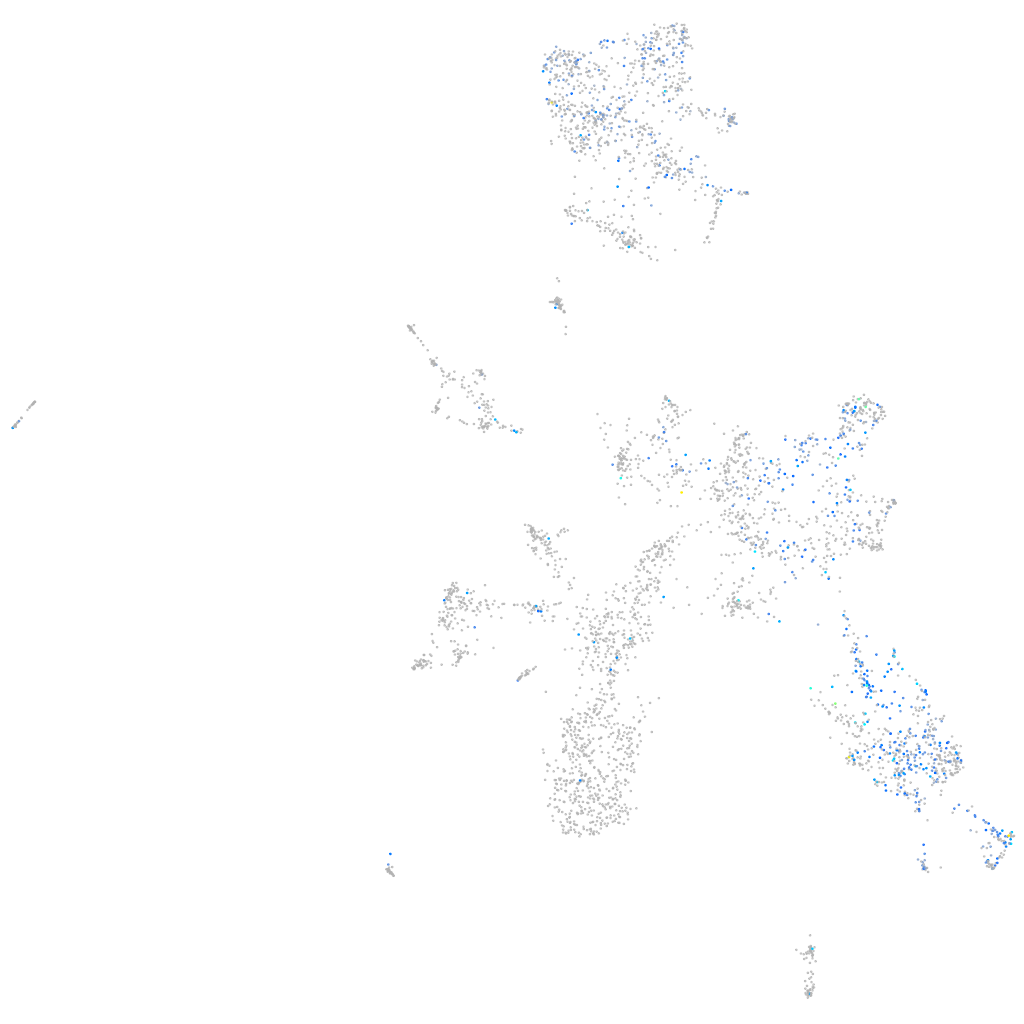
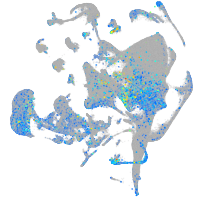
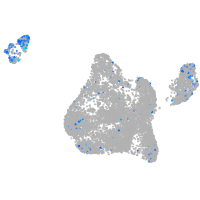
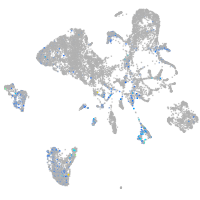
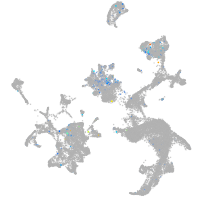
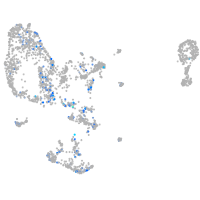
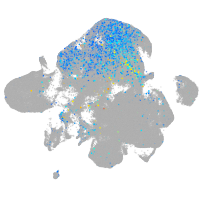
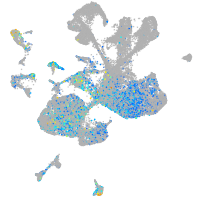
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| krt18b | 0.321 | elavl3 | -0.130 |
| si:ch211-243g18.2 | 0.320 | rtn1a | -0.127 |
| elf3 | 0.308 | insm1a | -0.123 |
| krt8 | 0.290 | atp6v0cb | -0.120 |
| s100a10b | 0.278 | insm1b | -0.110 |
| cnn2 | 0.277 | tmeff1b | -0.099 |
| cldne | 0.277 | ebf3a | -0.098 |
| zgc:158343 | 0.271 | sox11a | -0.098 |
| krt1-c5 | 0.266 | stmn1b | -0.096 |
| LOC799574 | 0.264 | tuba1c | -0.095 |
| malb | 0.262 | hmgb3a | -0.093 |
| her6 | 0.258 | zgc:65894 | -0.092 |
| wu:fj16a03 | 0.254 | neurod1 | -0.092 |
| capn8 | 0.253 | vamp2 | -0.091 |
| cfl1l | 0.248 | epb41a | -0.091 |
| si:dkey-247k7.2 | 0.247 | nova2 | -0.089 |
| tmem176l.4 | 0.246 | scg2a | -0.089 |
| zgc:174938 | 0.242 | rnasekb | -0.088 |
| dnase1l4.1 | 0.236 | atp6v1e1b | -0.087 |
| zfp36l1a | 0.236 | CR383676.1 | -0.087 |
| her9 | 0.236 | lhx2a | -0.085 |
| krt4 | 0.234 | gng3 | -0.085 |
| sult2st3 | 0.231 | tuba1a | -0.084 |
| sp8a | 0.231 | vdac3 | -0.083 |
| gsta.1 | 0.230 | si:ch211-137a8.4 | -0.083 |
| zgc:101785 | 0.229 | cnrip1a | -0.083 |
| krt18a.1 | 0.228 | sncb | -0.083 |
| lgals3b | 0.228 | si:dkeyp-75h12.5 | -0.082 |
| ftr83 | 0.226 | myt1a | -0.082 |
| cebpb | 0.225 | calm1a | -0.081 |
| tspan13a | 0.223 | calm2a | -0.081 |
| si:dkeyp-86h10.3 | 0.222 | ebf2 | -0.080 |
| pycard | 0.222 | dnajc5aa | -0.080 |
| klf6a | 0.220 | stxbp1a | -0.080 |
| si:ch73-31d8.2 | 0.219 | gsna | -0.079 |