LIM domain and actin binding 1a
ZFIN 
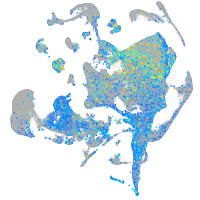
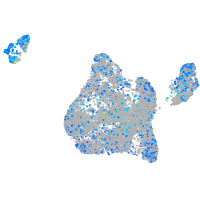
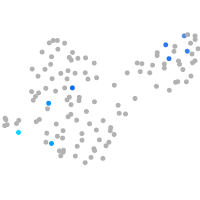
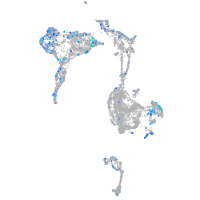
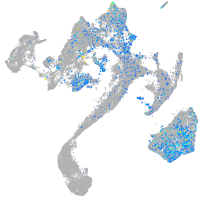
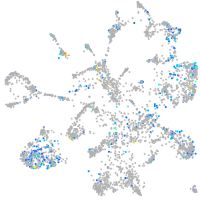
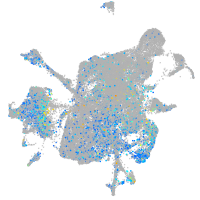
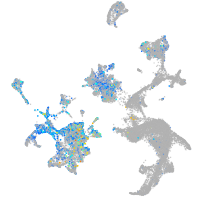
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-23i12.7 | 0.187 | gpx2 | -0.115 |
| eef1a1l1 | 0.164 | tspan13b | -0.114 |
| eif4ebp3l | 0.157 | si:ch211-243g18.2 | -0.112 |
| marcksl1b | 0.156 | osbpl1a | -0.111 |
| cirbpb | 0.156 | dnajc5b | -0.111 |
| ier5l | 0.152 | spa17 | -0.110 |
| khdrbs1a | 0.151 | atp1a3b | -0.109 |
| hnrnpa0l | 0.151 | si:ch73-15n15.3 | -0.108 |
| sfrp2 | 0.149 | lrrc73 | -0.108 |
| klf6a | 0.148 | COX3 | -0.108 |
| zfp36l1a | 0.147 | cd164l2 | -0.107 |
| cited4b | 0.145 | calml4a | -0.107 |
| si:ch211-222l21.1 | 0.142 | pou4f3 | -0.105 |
| atf3 | 0.142 | CR925719.1 | -0.105 |
| XLOC-042222 | 0.141 | otofb | -0.105 |
| s1pr5a | 0.139 | atp2b1a | -0.105 |
| tpm3 | 0.138 | nptna | -0.103 |
| rps3a | 0.137 | zgc:162989 | -0.103 |
| krt8 | 0.136 | emb | -0.103 |
| abracl | 0.136 | eno1a | -0.103 |
| mycn | 0.136 | grxcr2 | -0.102 |
| marcksl1a | 0.136 | gipc3 | -0.102 |
| ier5 | 0.135 | gpx1b | -0.102 |
| actb2 | 0.134 | apba1a | -0.102 |
| rsl24d1 | 0.132 | mt-atp6 | -0.100 |
| nppc | 0.132 | gb:eh456644 | -0.100 |
| zbtb16a | 0.130 | atp1b2b | -0.099 |
| h3f3d | 0.130 | tmprss3a | -0.099 |
| id1 | 0.129 | rbm24a | -0.099 |
| ptmaa | 0.128 | slc17a8 | -0.098 |
| rpl7a | 0.128 | gfi1aa | -0.098 |
| naca | 0.127 | vsig10l2 | -0.097 |
| CABZ01075068.1 | 0.127 | myo6b | -0.097 |
| hnrnpaba | 0.126 | eef1a1b | -0.097 |
| tmsb4x | 0.125 | tmc2a | -0.097 |