"ligase I, DNA, ATP-dependent"
ZFIN 
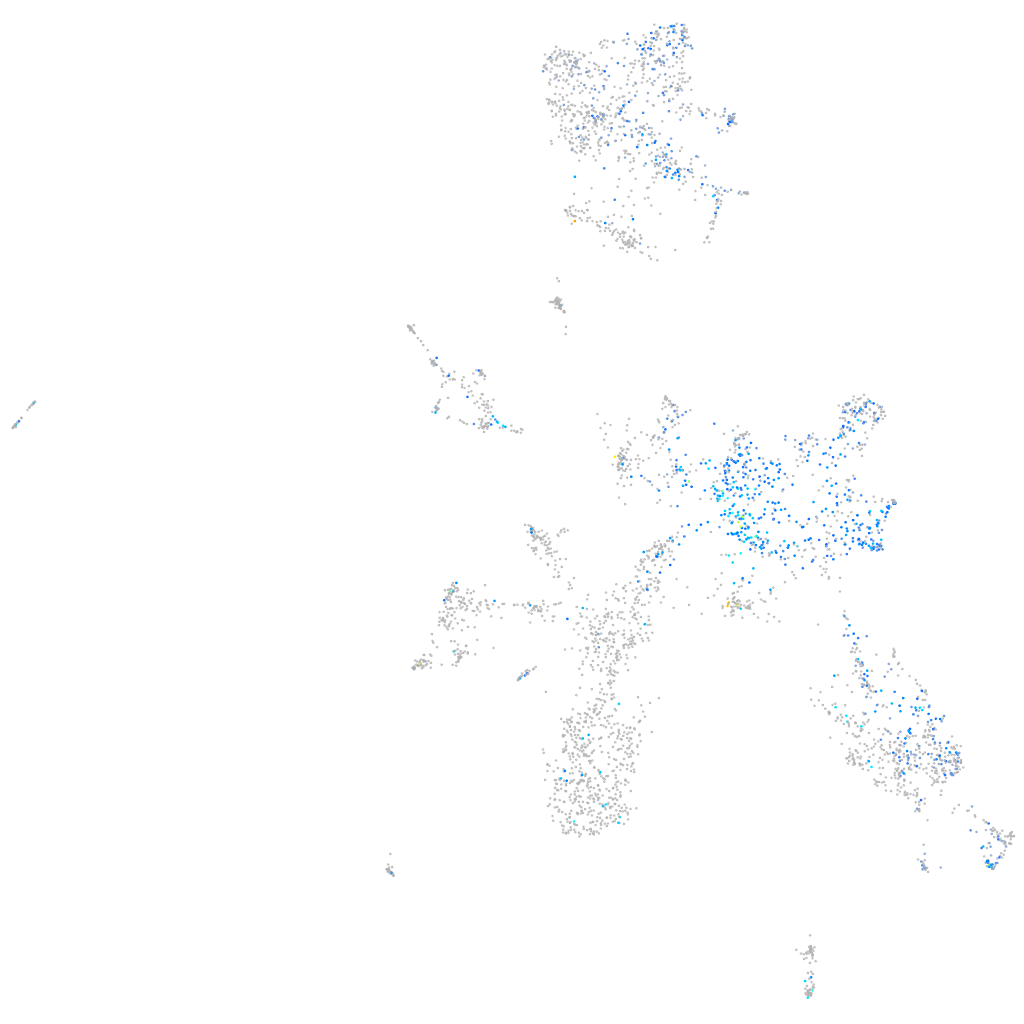
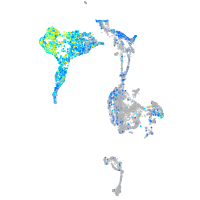
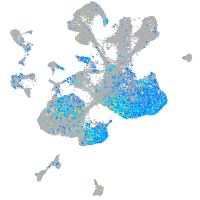
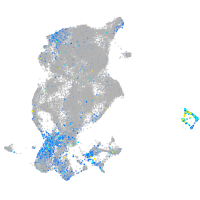
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.554 | txn | -0.285 |
| pcna | 0.537 | anxa5b | -0.279 |
| chaf1a | 0.534 | cldnh | -0.251 |
| rpa2 | 0.515 | gstp1 | -0.251 |
| rpa3 | 0.510 | tmem59 | -0.238 |
| rrm1 | 0.508 | CR383676.1 | -0.233 |
| mcm7 | 0.508 | gapdhs | -0.233 |
| nasp | 0.506 | calm1a | -0.231 |
| zgc:110540 | 0.506 | rtn1a | -0.225 |
| slbp | 0.500 | ccni | -0.216 |
| fen1 | 0.498 | selenow2b | -0.216 |
| rrm2 | 0.495 | rnasekb | -0.216 |
| stmn1a | 0.494 | bik | -0.215 |
| CABZ01005379.1 | 0.493 | fxyd1 | -0.214 |
| esco2 | 0.469 | gabarapa | -0.209 |
| mibp | 0.452 | vamp2 | -0.208 |
| tk1 | 0.449 | gnb1a | -0.208 |
| si:ch211-156b7.4 | 0.438 | atp6v1g1 | -0.208 |
| mcm4 | 0.436 | atp6v1e1b | -0.205 |
| rfc4 | 0.436 | selenow1 | -0.205 |
| prim2 | 0.429 | atp6v0cb | -0.203 |
| dhfr | 0.421 | chga | -0.200 |
| her2 | 0.418 | pvalb5 | -0.197 |
| orc4 | 0.415 | itm2ba | -0.196 |
| shmt1 | 0.413 | camk2n1a | -0.195 |
| asf1ba | 0.412 | wu:fb18f06 | -0.195 |
| zgc:110216 | 0.406 | smdt1b | -0.193 |
| LOC100330864 | 0.406 | nupr1a | -0.192 |
| nutf2l | 0.406 | ip6k2a | -0.192 |
| suv39h1b | 0.405 | nfe2l2a | -0.191 |
| fbxo5 | 0.404 | zgc:65894 | -0.191 |
| mcm6 | 0.404 | gabarapb | -0.191 |
| gins1 | 0.404 | f11r.1 | -0.190 |
| ranbp1 | 0.397 | rab6bb | -0.190 |
| tyms | 0.396 | tspan2a | -0.187 |