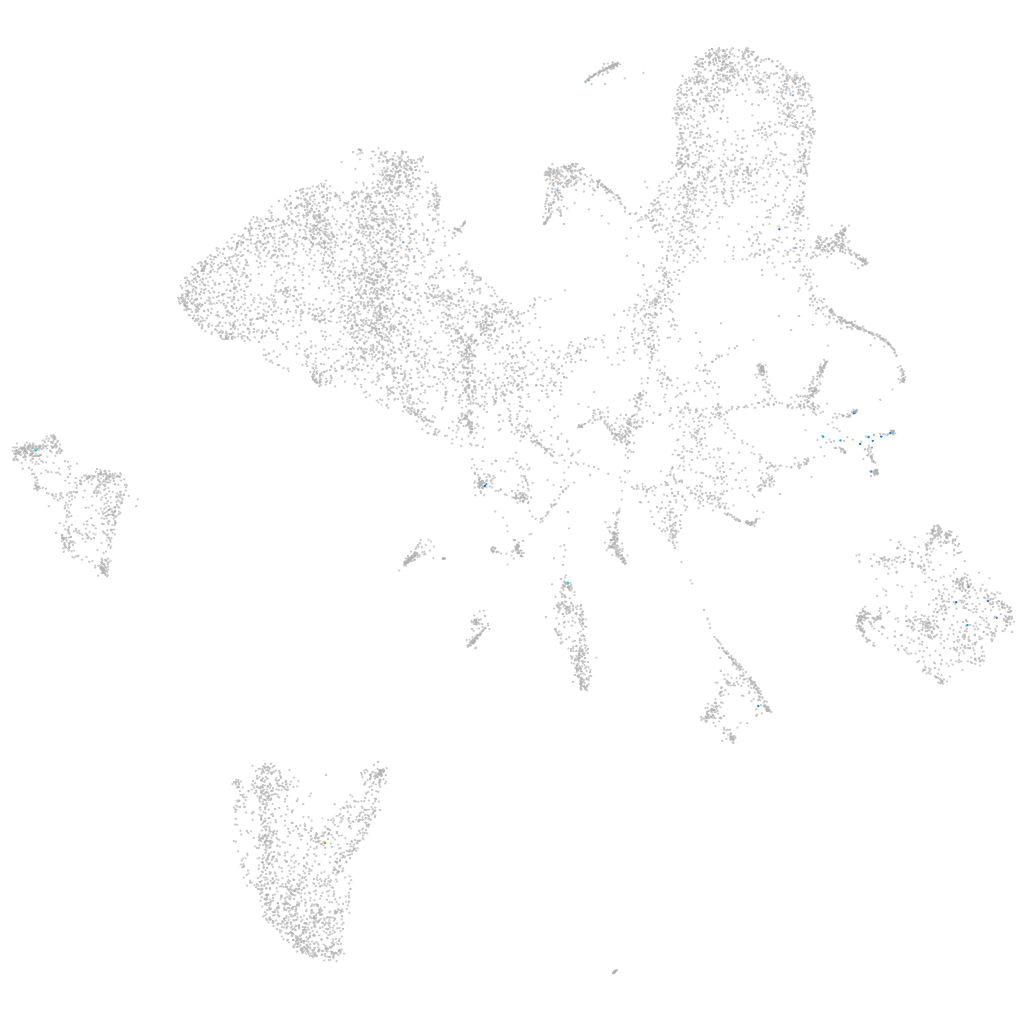
LHFPL tetraspan subfamily member 4b
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gnrhr1 | 0.487 | rpl10a | -0.046 |
| cul5b | 0.455 | gapdh | -0.046 |
| CR855860.1 | 0.340 | eef1da | -0.044 |
| nlgn3a | 0.327 | rps12 | -0.043 |
| oprk1 | 0.303 | suclg1 | -0.041 |
| XLOC-005794 | 0.302 | rpl5a | -0.040 |
| ntrk2a | 0.284 | nupr1b | -0.040 |
| hcar1-3 | 0.253 | cox6b1 | -0.040 |
| ppfia4 | 0.253 | tuba8l2 | -0.039 |
| tubb2 | 0.252 | rpl17 | -0.039 |
| fam131ba | 0.251 | aldob | -0.039 |
| CR786577.1 | 0.230 | glud1b | -0.039 |
| lhfpl3 | 0.218 | atp5l | -0.039 |
| cyp39a1 | 0.217 | gatm | -0.039 |
| XLOC-039021 | 0.211 | gstp1 | -0.039 |
| neto2a | 0.204 | aldh6a1 | -0.039 |
| BX248521.1 | 0.190 | mt-nd1 | -0.038 |
| CU914760.1 | 0.190 | gamt | -0.038 |
| prf1.3 | 0.190 | fbp1b | -0.037 |
| cplx2l | 0.185 | rplp1 | -0.037 |
| atp1a2a | 0.184 | eef1b2 | -0.037 |
| pimr189 | 0.182 | rpl32 | -0.036 |
| BX005245.1 | 0.181 | rps15 | -0.036 |
| NPAS3 | 0.180 | gsta.1 | -0.036 |
| CABZ01044764.2 | 0.179 | vdac2 | -0.036 |
| march9 | 0.177 | cx32.3 | -0.035 |
| adgrb1a | 0.175 | eno3 | -0.035 |
| edil3a | 0.172 | rpl14 | -0.035 |
| sema4aa | 0.170 | gpx1a | -0.035 |
| slc17a6a | 0.169 | mat1a | -0.035 |
| fli1b | 0.165 | rps3a | -0.035 |
| FP017274.1 | 0.164 | BX908782.3 | -0.034 |
| luzp2 | 0.158 | ndufa4l | -0.034 |
| gabrg2 | 0.156 | mt-atp6 | -0.034 |
| BX927369.1 | 0.155 | gpx4a | -0.034 |