"lectin, galactoside binding soluble 3a"
ZFIN 
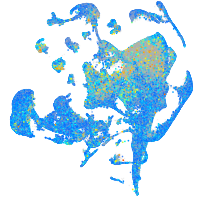
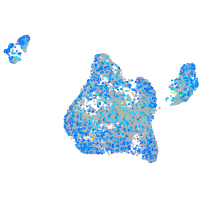
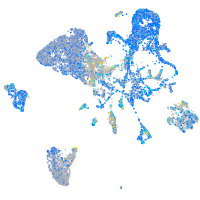
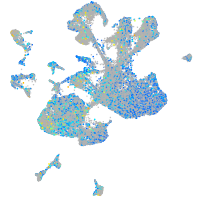
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cd9b | 0.341 | dkc1 | -0.204 |
| anxa4 | 0.317 | gnl3 | -0.194 |
| ndrg3a | 0.297 | cdx4 | -0.194 |
| slc25a5 | 0.294 | ncl | -0.193 |
| cldnc | 0.291 | apoc1 | -0.191 |
| krt8 | 0.291 | marcksl1b | -0.190 |
| krt18a.1 | 0.290 | nop58 | -0.189 |
| prkab1b | 0.287 | marcksb | -0.188 |
| epcam | 0.277 | ddx18 | -0.184 |
| si:dkey-112a7.4 | 0.269 | wu:fb97g03 | -0.183 |
| atp5meb | 0.268 | fbl | -0.178 |
| vdac2 | 0.266 | NC-002333.4 | -0.178 |
| cldnb | 0.264 | npm1a | -0.176 |
| GCA | 0.264 | ebna1bp2 | -0.175 |
| cdh17 | 0.259 | bms1 | -0.172 |
| cldn7b | 0.258 | rbmx2 | -0.165 |
| cldnh | 0.258 | apoeb | -0.165 |
| atp1b1a | 0.258 | zmp:0000000624 | -0.165 |
| si:ch1073-429i10.3.1 | 0.257 | cxcl12a | -0.165 |
| ap1s1 | 0.255 | seta | -0.163 |
| vdac3 | 0.253 | rsl1d1 | -0.162 |
| etnk1 | 0.252 | snrnp70 | -0.162 |
| mdh2 | 0.252 | lig1 | -0.158 |
| spint2 | 0.251 | mphosph10 | -0.154 |
| si:ch211-195e19.1 | 0.251 | znfl2a | -0.154 |
| rps10 | 0.249 | ppig | -0.153 |
| atp5if1b | 0.244 | plk3 | -0.152 |
| rps17 | 0.244 | rrp1 | -0.152 |
| mycbp | 0.244 | mybbp1a | -0.152 |
| tmx2b | 0.242 | nop56 | -0.151 |
| si:dkey-16l2.17 | 0.242 | parp1 | -0.151 |
| idh2 | 0.241 | hspb1 | -0.151 |
| slc25a3b | 0.241 | tal1 | -0.150 |
| rgcc | 0.240 | hnrnpa1a | -0.150 |
| id2a | 0.239 | rps18 | -0.150 |