"lectin, galactoside-binding, soluble, 2a"
ZFIN 
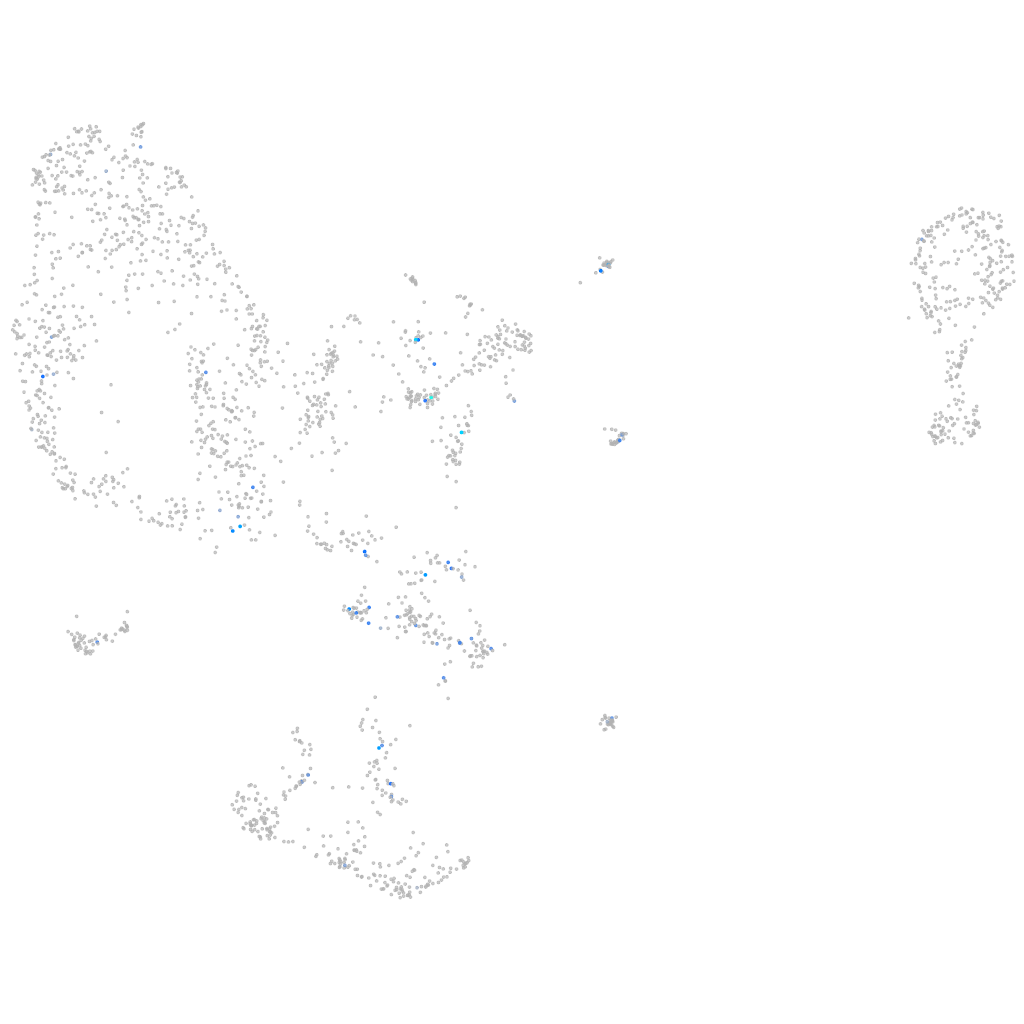
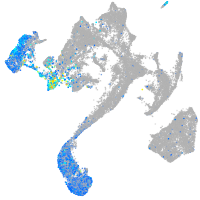
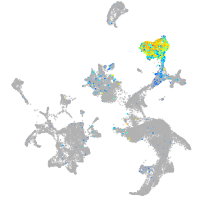
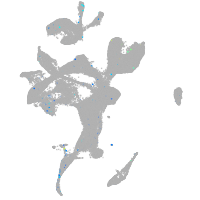
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| c1qa | 0.353 | gstt1a | -0.088 |
| myrf | 0.326 | atp5md | -0.087 |
| adra1ab | 0.317 | aldob | -0.085 |
| stard15 | 0.311 | hao1 | -0.085 |
| CU467822.2 | 0.291 | mt-nd3 | -0.085 |
| marco | 0.291 | eno3 | -0.084 |
| zgc:92162 | 0.291 | gpd1b | -0.083 |
| tbxa2r | 0.286 | pklr | -0.082 |
| XLOC-010278 | 0.282 | gstr | -0.082 |
| cxcl8b.1 | 0.280 | adka | -0.080 |
| CABZ01064941.1 | 0.273 | si:dkey-16p21.8 | -0.079 |
| mb21d2 | 0.264 | cubn | -0.078 |
| tnfaip6 | 0.262 | nipsnap3a | -0.078 |
| ankrd22 | 0.259 | fbp1b | -0.078 |
| crfb16 | 0.255 | dpys | -0.078 |
| dip2bb | 0.255 | glud1b | -0.077 |
| mir203a | 0.251 | zgc:136493 | -0.077 |
| CR318601.1 | 0.241 | agxta | -0.076 |
| CT737123.1 | 0.241 | dab2 | -0.075 |
| pdyn | 0.241 | eps8l3a | -0.074 |
| mpp4a | 0.240 | rps18 | -0.074 |
| ifitm1 | 0.236 | hoga1 | -0.073 |
| itgae.1 | 0.232 | gapdh | -0.073 |
| XLOC-020301 | 0.232 | cox7a1 | -0.072 |
| cmklr1 | 0.229 | grhprb | -0.072 |
| si:ch211-197l9.2 | 0.227 | aldh7a1 | -0.072 |
| BX004996.1 | 0.227 | lrp2a | -0.072 |
| CU570682.1 | 0.220 | cox6a2 | -0.071 |
| chst8 | 0.218 | aco1 | -0.071 |
| si:dkeyp-35e5.5 | 0.218 | alpl | -0.071 |
| ugt2a2 | 0.218 | dera | -0.070 |
| FP236741.1 | 0.218 | upb1 | -0.069 |
| LO018196.1 | 0.216 | si:dkey-28n18.9 | -0.069 |
| chia.2 | 0.212 | suclg2 | -0.069 |
| si:dkey-63j12.4 | 0.212 | mt-co1 | -0.069 |