lactate dehydrogenase D
ZFIN 
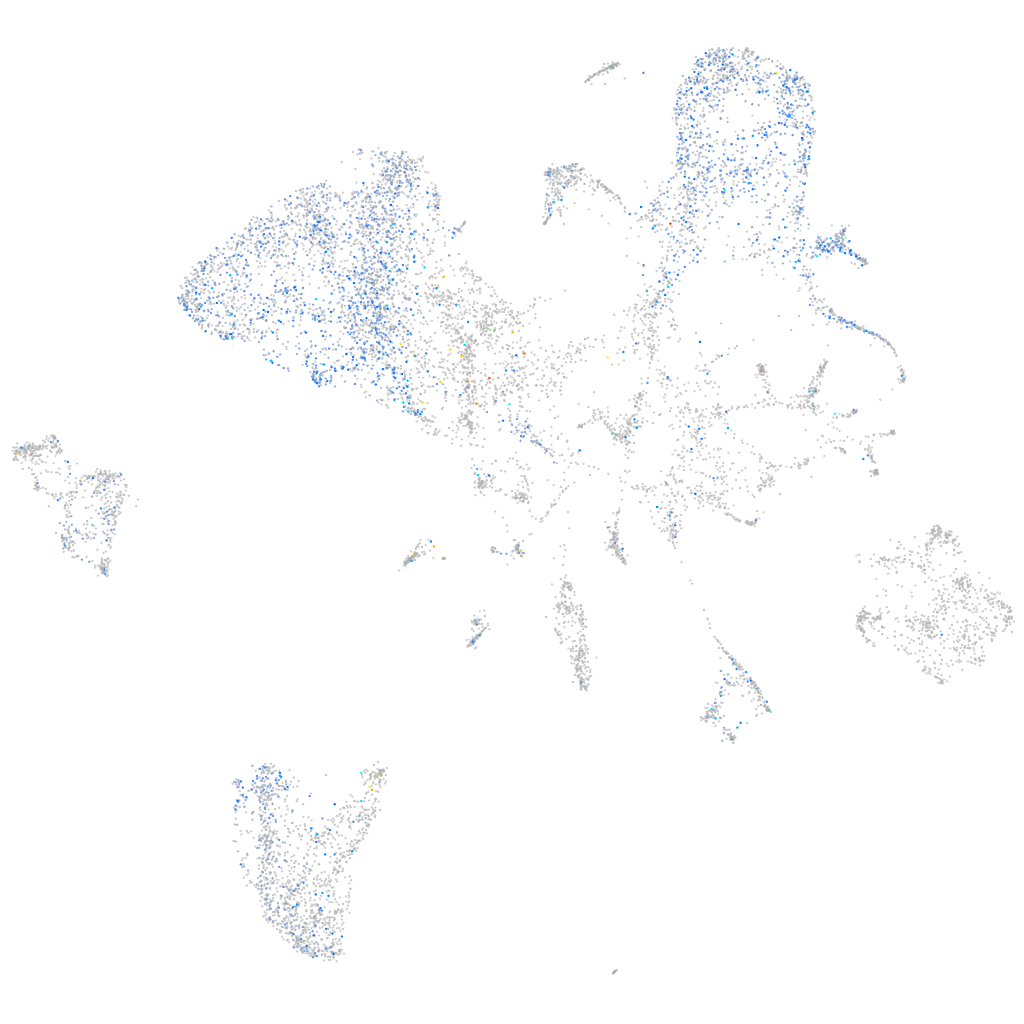
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| glud1b | 0.238 | marcksb | -0.140 |
| ugt1a7 | 0.227 | marcksl1b | -0.139 |
| srd5a2a | 0.227 | hmgb3a | -0.139 |
| sod1 | 0.226 | hmgn6 | -0.128 |
| slc37a4a | 0.225 | hspb1 | -0.127 |
| atp5f1b | 0.225 | cx43.4 | -0.120 |
| sult2st2 | 0.223 | akap12b | -0.117 |
| atp5mc1 | 0.221 | hmgb1b | -0.116 |
| eno3 | 0.220 | tgif1 | -0.116 |
| rdh1 | 0.219 | jpt1b | -0.113 |
| atp5fa1 | 0.219 | si:ch211-152c2.3 | -0.113 |
| scp2a | 0.216 | h3f3d | -0.112 |
| mt-nd1 | 0.214 | si:ch211-222l21.1 | -0.111 |
| mt-atp6 | 0.214 | cdh6 | -0.109 |
| cyp8b1 | 0.213 | glulb | -0.109 |
| dhrs9 | 0.212 | hnrnpa0a | -0.109 |
| gstk1 | 0.211 | nucks1a | -0.108 |
| mdh1aa | 0.211 | si:ch211-137a8.4 | -0.108 |
| gapdh | 0.210 | si:ch73-1a9.3 | -0.107 |
| mttp | 0.209 | smarcb1b | -0.105 |
| acadm | 0.209 | syncrip | -0.102 |
| fdx1 | 0.208 | hnrnpaba | -0.102 |
| gstt1a | 0.208 | id1 | -0.101 |
| suclg1 | 0.207 | hes6 | -0.101 |
| cx32.3 | 0.205 | hdac1 | -0.098 |
| fbp1b | 0.204 | zgc:110425 | -0.097 |
| sdr16c5b | 0.203 | aldoaa | -0.097 |
| haao | 0.203 | tpm4a | -0.096 |
| tecrb | 0.203 | apoeb | -0.096 |
| si:ch211-107o10.3 | 0.203 | eef1a1l1 | -0.095 |
| chchd10 | 0.203 | tspan7 | -0.095 |
| gstr | 0.202 | ube2e1 | -0.095 |
| pgk1 | 0.202 | prtfdc1 | -0.094 |
| acsf2 | 0.200 | hmgb1a | -0.094 |
| acadl | 0.200 | qkia | -0.094 |