LIM domain binding 2a
ZFIN 
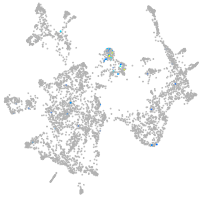
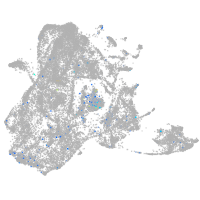
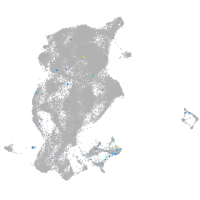
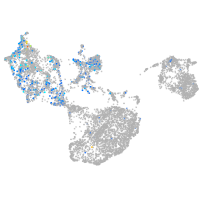
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pecam1 | 0.504 | c1galt1c1 | -0.042 |
| gja5a | 0.492 | slc25a55a | -0.040 |
| sez6b | 0.485 | capn2b | -0.040 |
| flt1 | 0.480 | ak2 | -0.039 |
| ecscr | 0.462 | tspan15 | -0.039 |
| myoz2b | 0.447 | si:ch73-359m17.9 | -0.039 |
| zgc:158423 | 0.433 | cldnh | -0.038 |
| si:ch211-191c10.1 | 0.430 | zgc:110339 | -0.036 |
| fam129ab | 0.421 | b3gnt5a | -0.035 |
| hyal2a | 0.414 | ciapin1 | -0.035 |
| ankrd6a | 0.378 | jupa | -0.034 |
| PITPNC1 | 0.369 | tmem54a | -0.034 |
| si:rp71-1c10.8 | 0.369 | cldn7b | -0.034 |
| LOC110439847 | 0.369 | ethe1 | -0.034 |
| myct1b | 0.365 | LOC799574 | -0.034 |
| nrn1lb | 0.364 | capn9 | -0.033 |
| lpl | 0.360 | mat2al | -0.033 |
| LOC101885341 | 0.357 | si:dkey-87o1.2 | -0.033 |
| si:ch211-214c7.5 | 0.353 | rpl5a | -0.033 |
| f7i | 0.343 | naga | -0.033 |
| trpm1a | 0.338 | cpeb4b | -0.033 |
| nt5c1aa | 0.337 | dnase1l4.1 | -0.033 |
| BX663503.1 | 0.337 | gata3 | -0.033 |
| bcl6b | 0.327 | zgc:110333 | -0.033 |
| slc1a7a | 0.325 | myzap | -0.033 |
| thegl | 0.319 | gfpt2 | -0.032 |
| glipr2l | 0.318 | sgk2a | -0.032 |
| rasip1 | 0.316 | ddx21 | -0.032 |
| rgs16 | 0.313 | itm2bb | -0.032 |
| si:ch211-14k19.8 | 0.311 | lpar2b | -0.032 |
| pald1a | 0.308 | chmp2bb | -0.031 |
| calb2b | 0.308 | ndrg1a | -0.031 |
| thsd1 | 0.308 | rpl38 | -0.031 |
| sult4a1 | 0.305 | tollip | -0.031 |
| nr1d4a | 0.301 | glod5 | -0.031 |