lymphocyte cytosolic protein 1 (L-plastin)
ZFIN 
Other cell groups


all cells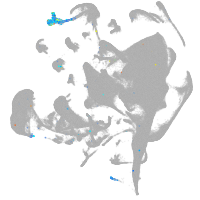 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 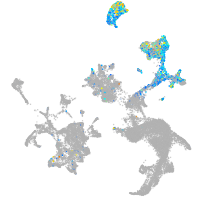 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| coro1a | 0.609 | hbbe1.3 | -0.292 |
| arpc1b | 0.573 | hbae1.1 | -0.292 |
| fcer1gl | 0.553 | hbae3 | -0.291 |
| rac2 | 0.553 | hbae1.3 | -0.287 |
| laptm5 | 0.550 | hbbe2 | -0.285 |
| ctss2.1 | 0.537 | hbbe1.1 | -0.276 |
| spi1b | 0.529 | cahz | -0.263 |
| pfn1 | 0.529 | hbbe1.2 | -0.249 |
| cebpb | 0.526 | fth1a | -0.246 |
| cotl1 | 0.514 | hemgn | -0.245 |
| glipr1a | 0.506 | alas2 | -0.231 |
| hcls1 | 0.500 | blvrb | -0.230 |
| si:dkey-102g19.3 | 0.493 | si:ch211-250g4.3 | -0.228 |
| cyba | 0.492 | zgc:56095 | -0.227 |
| ptpn6 | 0.491 | creg1 | -0.222 |
| si:ch211-102c2.4 | 0.488 | lmo2 | -0.216 |
| zgc:64051 | 0.473 | slc4a1a | -0.216 |
| il1b | 0.471 | nt5c2l1 | -0.212 |
| skap2 | 0.471 | zgc:163057 | -0.203 |
| lta4h | 0.469 | epb41b | -0.202 |
| arhgdig | 0.468 | si:ch211-207c6.2 | -0.195 |
| arpc5b | 0.464 | nmt1b | -0.184 |
| grap2b | 0.463 | aqp1a.1 | -0.179 |
| capgb | 0.462 | tmod4 | -0.179 |
| itgb2 | 0.455 | tspo | -0.177 |
| wasb | 0.455 | plac8l1 | -0.169 |
| fam49ba | 0.452 | hbae5 | -0.165 |
| ptprc | 0.451 | si:ch211-227m13.1 | -0.164 |
| eno3 | 0.446 | uros | -0.162 |
| ncf1 | 0.444 | sptb | -0.161 |
| FERMT3 (1 of many) | 0.440 | tfr1a | -0.157 |
| alox5ap | 0.438 | hbbe3 | -0.154 |
| lect2l | 0.435 | rfesd | -0.153 |
| arpc4 | 0.433 | dmtn | -0.153 |
| mapre1a | 0.425 | arl4aa | -0.150 |




