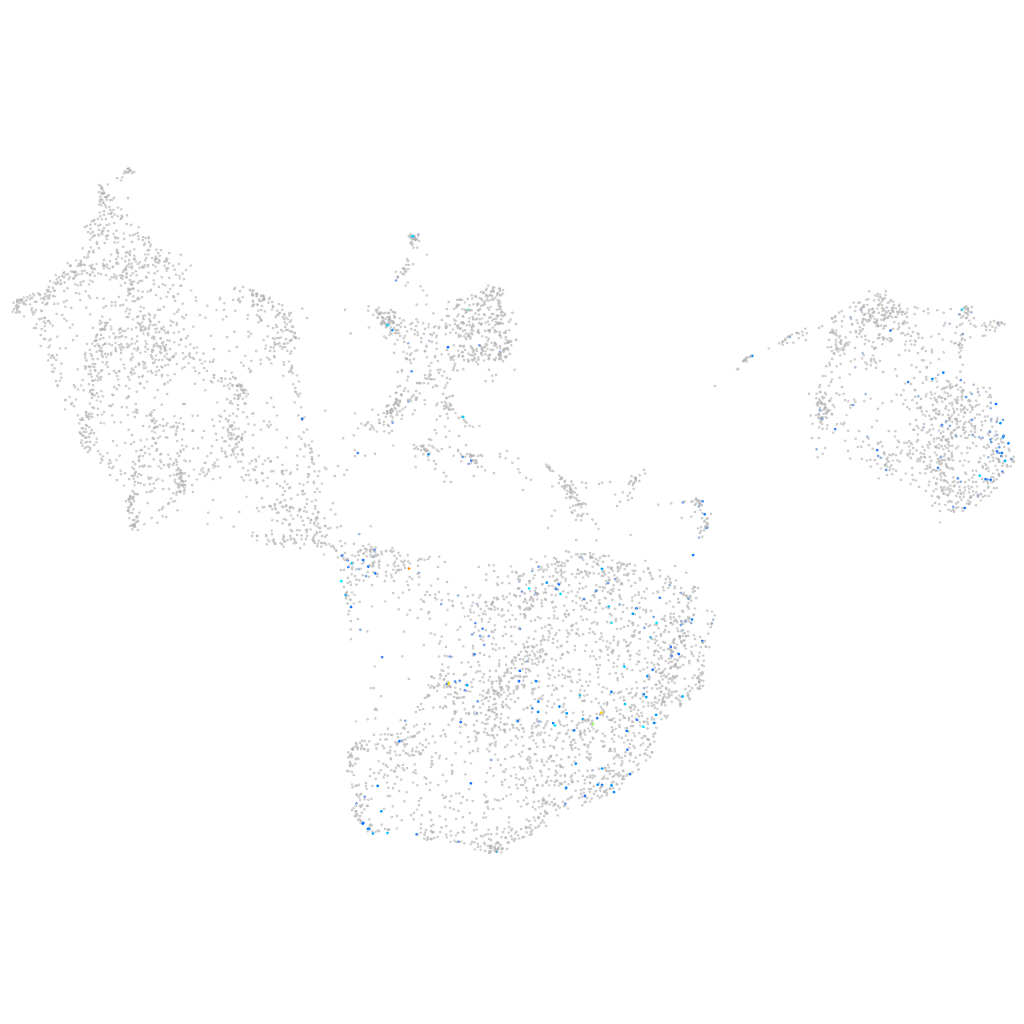
"laminin, gamma 2"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis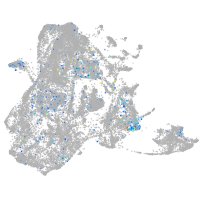 |
periderm |
fin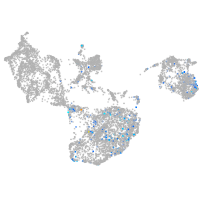 |
ionocytes / mucous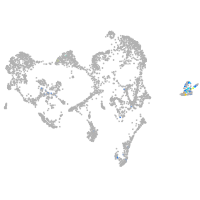 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| avpr2l | 0.236 | c1qtnf5 | -0.093 |
| XLOC-023402 | 0.198 | bhmt | -0.089 |
| LOC108183500 | 0.166 | fbln1 | -0.086 |
| si:cabz01036022.1 | 0.161 | hgd | -0.086 |
| LOC100536609 | 0.148 | marcksl1a | -0.086 |
| BX640512.3 | 0.146 | cfl1 | -0.082 |
| her5 | 0.145 | cd82a | -0.082 |
| si:ch73-193i2.2 | 0.143 | pcbd1 | -0.081 |
| ptgdsb.1 | 0.140 | twist1a | -0.080 |
| si:dkey-248g15.3 | 0.140 | ttc36 | -0.080 |
| tmem238a | 0.136 | tmem119b | -0.078 |
| znf1082 | 0.135 | qdpra | -0.075 |
| spaca4l | 0.134 | hpdb | -0.074 |
| s100a10b | 0.134 | ptx3a | -0.074 |
| anxa1a | 0.134 | gpc1a | -0.074 |
| itga6b | 0.133 | prrx1a | -0.073 |
| cldni | 0.133 | prrx1b | -0.073 |
| hbegfa | 0.131 | reck | -0.070 |
| epcam | 0.130 | fah | -0.069 |
| capgb | 0.129 | cd81a | -0.069 |
| cldn1 | 0.129 | tuba8l3 | -0.065 |
| jupa | 0.127 | slc16a10 | -0.065 |
| LOC101882663 | 0.127 | si:dkey-151g10.6 | -0.064 |
| cfl1l | 0.127 | hoxa13b | -0.064 |
| gstm.3 | 0.126 | pah | -0.064 |
| tmsb1 | 0.125 | twist3 | -0.064 |
| si:dkey-102c8.3 | 0.125 | dcn | -0.063 |
| zgc:92380 | 0.124 | crabp2a | -0.063 |
| apoeb | 0.123 | rpl13a | -0.062 |
| epgn | 0.122 | si:ch211-133n4.4 | -0.062 |
| si:ch211-249o11.5 | 0.122 | mdh1aa | -0.062 |
| tp63 | 0.121 | mfap2 | -0.062 |
| myh9a | 0.121 | twist1b | -0.061 |
| pfn1 | 0.121 | mxra5b | -0.061 |
| lgals3b | 0.121 | serpinf1 | -0.061 |