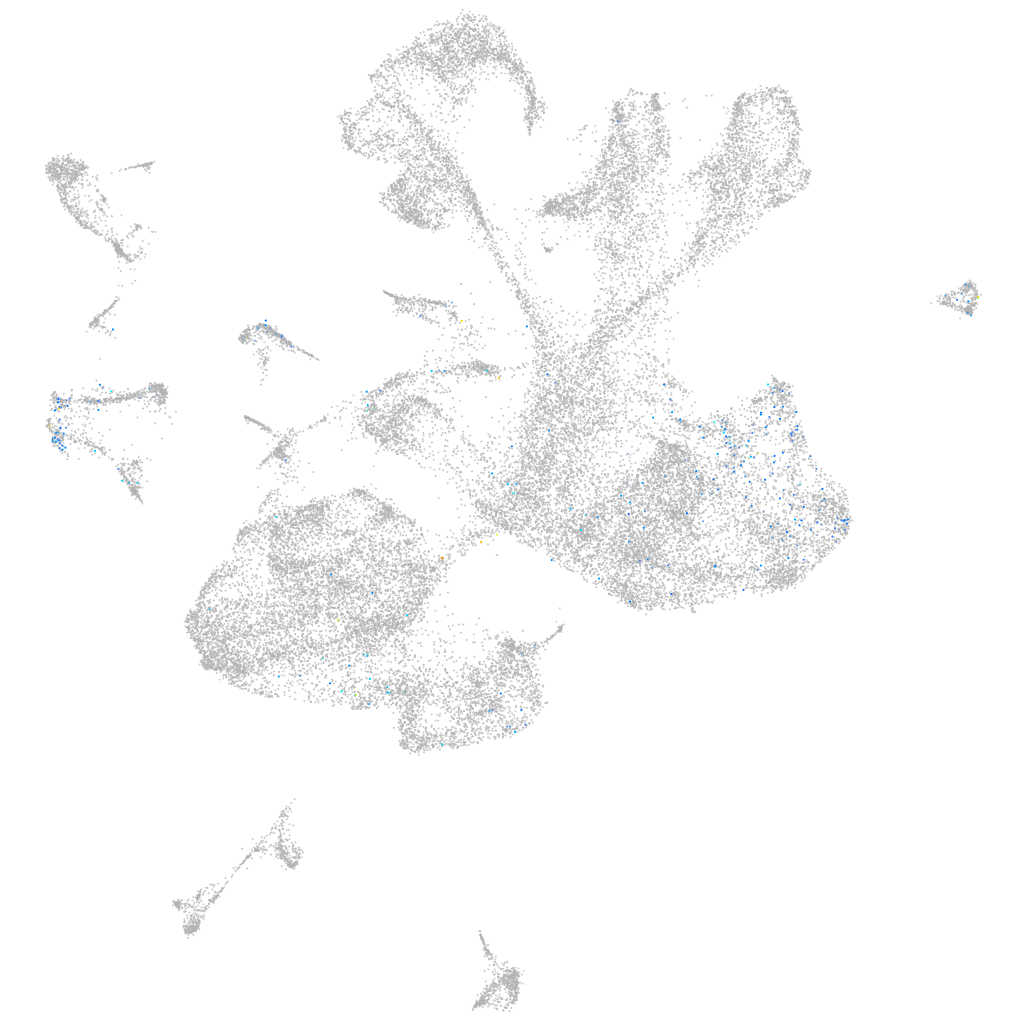
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.133 | marcksl1b | -0.046 |
| dnali1 | 0.127 | stmn1b | -0.044 |
| taar10 | 0.116 | tmsb4x | -0.043 |
| si:ch211-163l21.7 | 0.116 | rtn1a | -0.042 |
| ropn1l | 0.115 | elavl3 | -0.042 |
| si:dkey-148f10.4 | 0.111 | myt1b | -0.040 |
| ribc2 | 0.110 | ckbb | -0.039 |
| smkr1 | 0.109 | fabp3 | -0.039 |
| efcab1 | 0.108 | tmsb | -0.037 |
| enkur | 0.108 | hmgb3a | -0.034 |
| zgc:55461 | 0.107 | rnasekb | -0.033 |
| dnaaf4 | 0.106 | ppdpfb | -0.033 |
| pih1d3 | 0.105 | ywhah | -0.032 |
| ccdc151 | 0.103 | pvalb1 | -0.032 |
| ccdc40 | 0.102 | elavl4 | -0.032 |
| cfap45 | 0.101 | si:dkey-276j7.1 | -0.031 |
| tex9 | 0.100 | mdkb | -0.031 |
| rab36 | 0.100 | scrt2 | -0.031 |
| ccdc173 | 0.099 | nova1 | -0.031 |
| xrra1 | 0.098 | stx1b | -0.030 |
| rsph9 | 0.098 | si:dkeyp-75h12.5 | -0.030 |
| cfap126 | 0.097 | onecut1 | -0.030 |
| foxj1a | 0.095 | cadm3 | -0.030 |
| spag6 | 0.095 | rbfox1 | -0.030 |
| LOC108191721 | 0.095 | gng2 | -0.029 |
| meig1 | 0.094 | gng3 | -0.029 |
| ttc25 | 0.093 | si:ch73-386h18.1 | -0.028 |
| HTRA2 (1 of many).11 | 0.093 | gpm6ab | -0.028 |
| si:dkey-42p14.3 | 0.093 | thsd7aa | -0.028 |
| nme5 | 0.092 | onecut2 | -0.028 |
| spaca9 | 0.092 | nsg2 | -0.028 |
| cfap52 | 0.090 | zfhx3 | -0.028 |
| LOC101882848 | 0.089 | syt2a | -0.028 |
| rsph4a | 0.088 | rtn4b | -0.028 |
| cfap100 | 0.088 | atp6v0cb | -0.028 |