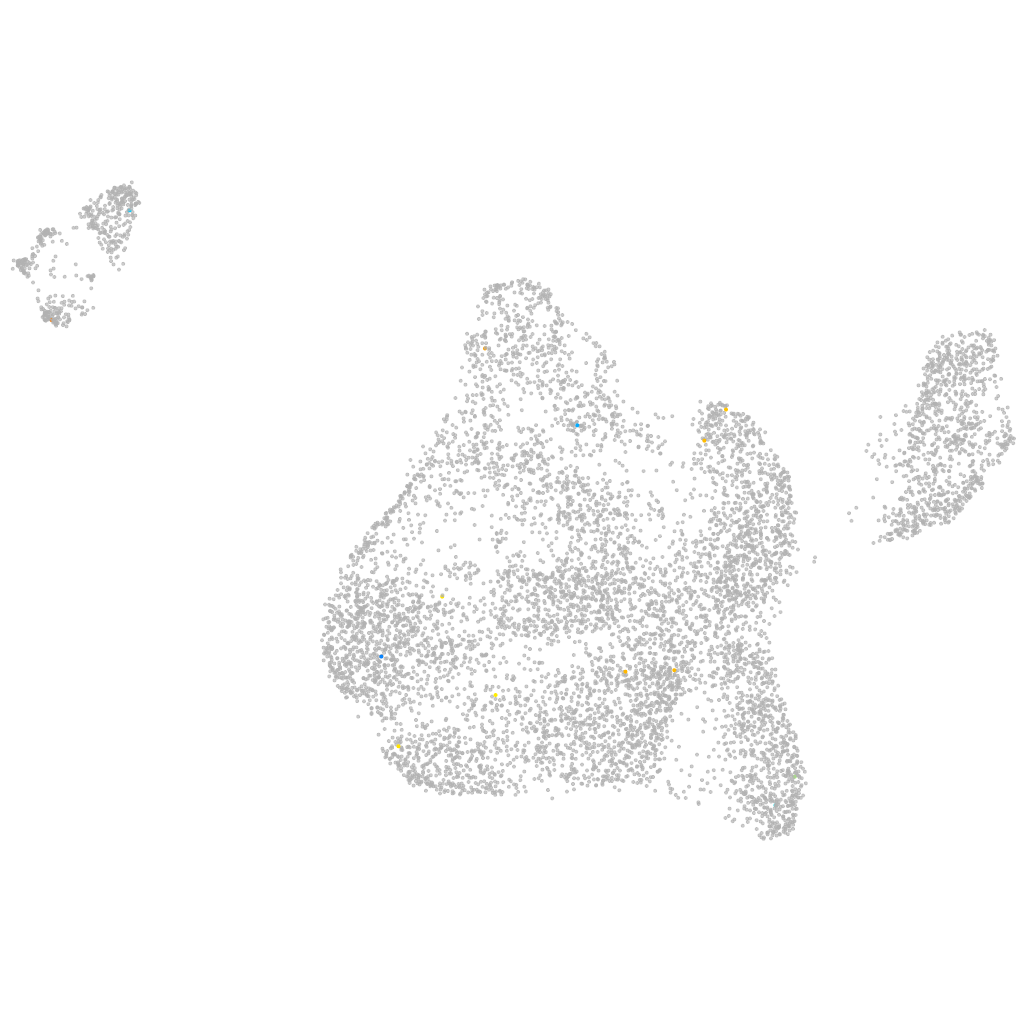
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-163g19.2 | 0.277 | chrac1 | -0.028 |
| pcdh1g30 | 0.270 | rps4x | -0.027 |
| lsp1 | 0.252 | prps1a | -0.026 |
| RF00093 | 0.242 | nsun2 | -0.025 |
| AL935184.2 | 0.236 | leo1 | -0.024 |
| XLOC-004886 | 0.195 | ddx52 | -0.024 |
| kcnj2a | 0.180 | nfyba | -0.023 |
| myo15ab | 0.170 | smc3 | -0.023 |
| CU861459.1 | 0.170 | nat9 | -0.023 |
| slc6a17 | 0.163 | add1 | -0.023 |
| BX537263.2 | 0.159 | gskip | -0.023 |
| dqx1 | 0.159 | exosc10 | -0.023 |
| XLOC-036261 | 0.159 | rpl24 | -0.023 |
| dicp1.16 | 0.151 | hspa14 | -0.023 |
| CT573429.2 | 0.146 | CABZ01087560.1 | -0.023 |
| XLOC-023279 | 0.140 | CR383676.1 | -0.023 |
| fgf1b | 0.130 | csnk1da | -0.022 |
| pkd1b | 0.127 | histh1l | -0.022 |
| smad3b | 0.125 | cdkn1ca | -0.022 |
| anxa2a | 0.122 | tp53inp2 | -0.022 |
| si:ch211-197f20.1 | 0.121 | DDX17 | -0.022 |
| ttll10 | 0.120 | cnbpa | -0.022 |
| apoa4b.2 | 0.120 | hnrnpm | -0.021 |
| ACVR1C | 0.117 | psma4 | -0.021 |
| itga2.2 | 0.114 | ccdc59 | -0.021 |
| CR749763.1 | 0.112 | spcs1 | -0.021 |
| agla | 0.111 | fip1l1b | -0.021 |
| hrh3 | 0.111 | sap30bp | -0.021 |
| stx7l | 0.111 | smarcc1a | -0.021 |
| fermt3b | 0.110 | ube2a | -0.021 |
| RF00158 | 0.107 | ddx5 | -0.021 |
| kcnd2 | 0.107 | mcm6 | -0.021 |
| CT573316.2 | 0.106 | pim1 | -0.020 |
| adam12 | 0.105 | hnrnpa1a | -0.020 |
| CU856344.2 | 0.101 | zc4h2 | -0.020 |