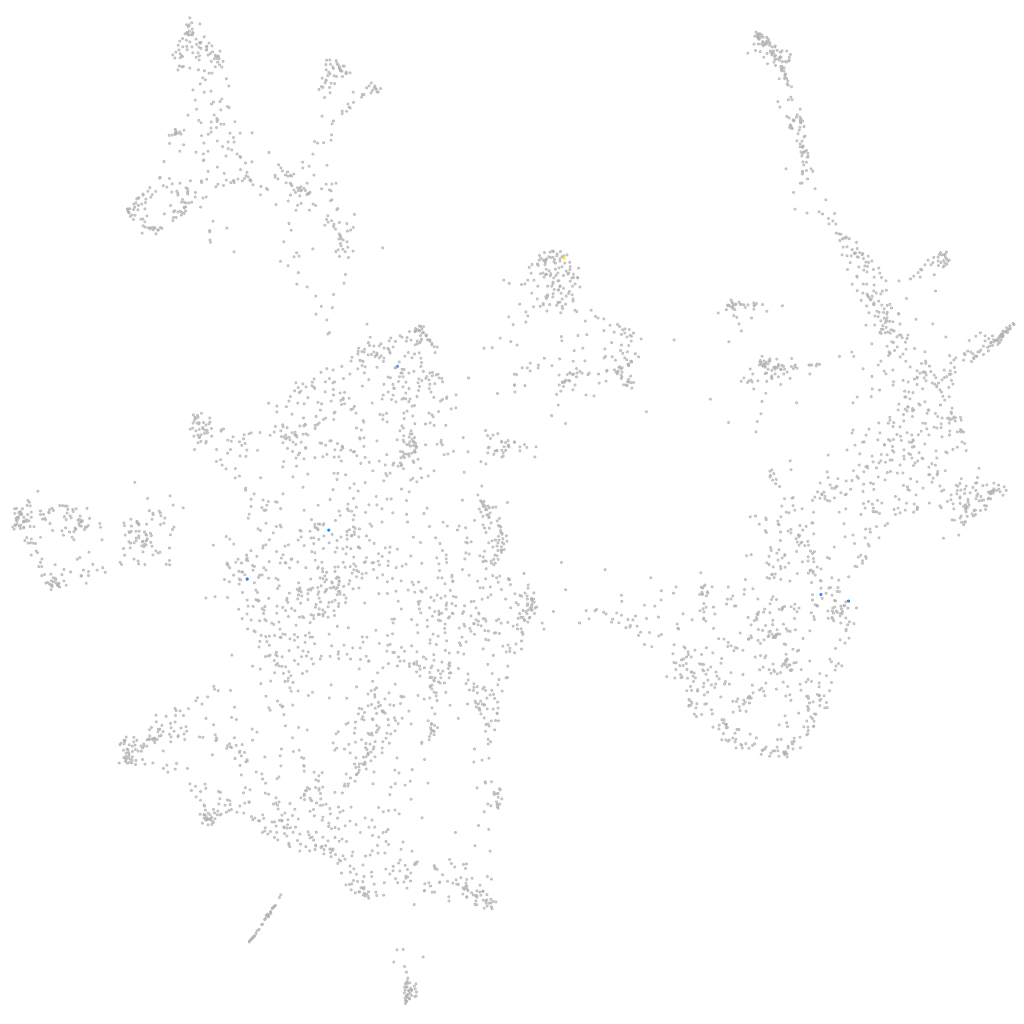
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR388166.1 | 0.824 | rps26l | -0.050 |
| zte38 | 0.770 | rpl10 | -0.050 |
| slc6a4b | 0.737 | rps5 | -0.045 |
| si:dkey-33c9.6 | 0.695 | rpl24 | -0.043 |
| fndc5a | 0.564 | eef1g | -0.039 |
| AL831726.1 | 0.544 | mt-nd1 | -0.036 |
| si:ch211-59d15.4 | 0.514 | ndufa4l | -0.033 |
| ppef1 | 0.511 | rpl23a | -0.033 |
| rap1gap2b | 0.503 | cox6b2 | -0.030 |
| XLOC-019059 | 0.502 | rpl31 | -0.027 |
| CU928046.1 | 0.483 | rpl32 | -0.026 |
| hrh3 | 0.483 | ctsla | -0.025 |
| gas2l2 | 0.472 | mt-nd4 | -0.025 |
| CT027980.1 | 0.454 | slc25a3b | -0.025 |
| kcng3 | 0.447 | cox7c | -0.024 |
| nkx2.4a | 0.428 | romo1 | -0.024 |
| CRACR2A | 0.427 | STMP1 | -0.024 |
| tmem121ab | 0.427 | eif3f | -0.024 |
| XLOC-026855 | 0.420 | ppiaa | -0.024 |
| calcr | 0.416 | ier2b | -0.024 |
| syngap1a | 0.410 | rps13 | -0.023 |
| tac1 | 0.398 | COX5B | -0.023 |
| cnn1b | 0.388 | si:dkey-16p21.8 | -0.022 |
| frs3 | 0.383 | cox8a | -0.022 |
| tnni1d | 0.383 | mdh2 | -0.021 |
| glis1a | 0.381 | atp5pb | -0.020 |
| dkk3a | 0.376 | ndufs6 | -0.020 |
| nkx2.4b | 0.369 | eef1da | -0.020 |
| si:ch211-122l14.7 | 0.367 | fip1l1b | -0.020 |
| CABZ01086574.1 | 0.365 | ctdsp2 | -0.020 |
| ccm2l | 0.357 | psmd4b | -0.020 |
| prdm13 | 0.357 | dap1b | -0.020 |
| TMEM114 | 0.350 | selenow1 | -0.020 |
| nkx2.2a | 0.349 | ccng1 | -0.020 |
| si:dkey-27j5.5 | 0.343 | etfb | -0.020 |