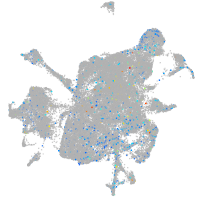
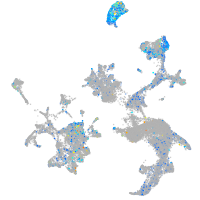
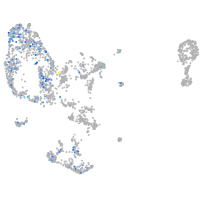
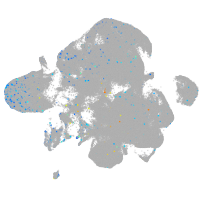
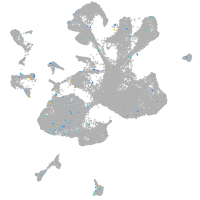
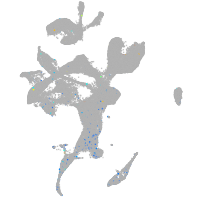
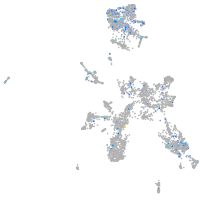
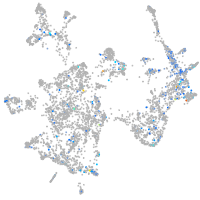
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elovl1b | 0.223 | si:ch73-281n10.2 | -0.156 |
| cubn | 0.211 | hmgb2b | -0.155 |
| slc26a1 | 0.204 | marcksb | -0.151 |
| aco1 | 0.199 | si:ch73-1a9.3 | -0.149 |
| mt-nd4 | 0.199 | hspb1 | -0.148 |
| oplah | 0.198 | hnrnpabb | -0.145 |
| glud1b | 0.197 | hmga1a | -0.144 |
| alpl | 0.196 | hmgb2a | -0.144 |
| ppifb | 0.195 | nucks1a | -0.142 |
| si:dkey-16p21.8 | 0.190 | wu:fb97g03 | -0.135 |
| mt-atp6 | 0.189 | h2afvb | -0.134 |
| abat | 0.189 | khdrbs1a | -0.133 |
| acsf2 | 0.188 | hmgb1b | -0.133 |
| aldh6a1 | 0.188 | setb | -0.131 |
| dmrt2b | 0.187 | h3f3d | -0.130 |
| mt-nd1 | 0.186 | marcksl1b | -0.130 |
| pdzk1ip1 | 0.186 | hmgn6 | -0.129 |
| lgmn | 0.186 | snrpe | -0.129 |
| nitr3a | 0.185 | cirbpb | -0.128 |
| kif5ab | 0.185 | syncrip | -0.125 |
| eno3 | 0.185 | hmgn2 | -0.125 |
| mt-nd2 | 0.183 | ptmab | -0.125 |
| alk | 0.183 | hnrnpaba | -0.124 |
| dpys | 0.183 | zgc:110425 | -0.121 |
| npr1b | 0.182 | ptges3b | -0.121 |
| bsg | 0.182 | apoeb | -0.121 |
| grhprb | 0.182 | seta | -0.120 |
| ctsla | 0.182 | cx43.4 | -0.119 |
| atp6v1e1b | 0.182 | ilf3b | -0.119 |
| enpp6 | 0.182 | hdac1 | -0.119 |
| CABZ01113467.1 | 0.181 | sumo3b | -0.119 |
| CU855854.1 | 0.181 | si:ch211-222l21.1 | -0.118 |
| utrnp | 0.181 | apoc1 | -0.118 |
| si:dkey-194e6.1 | 0.180 | hmgn7 | -0.115 |
| dpydb | 0.180 | si:ch211-288g17.3 | -0.115 |