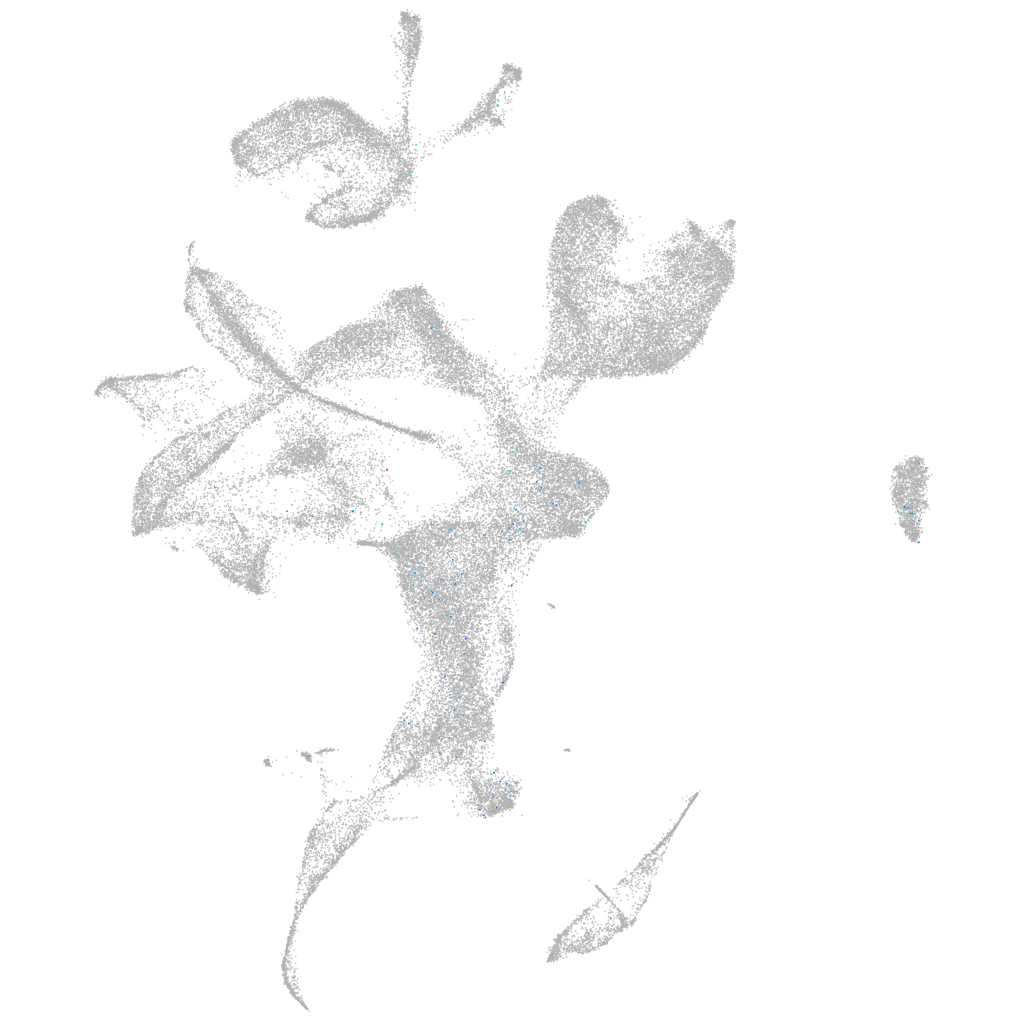
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 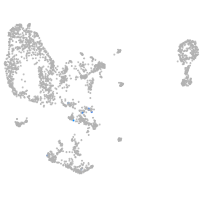 |
neural |
spinal cord / glia |
eye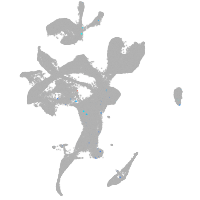 |
taste / olfactory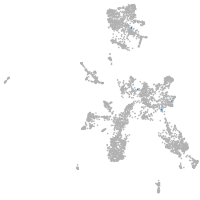 |
otic / lateral line |
epidermis |
periderm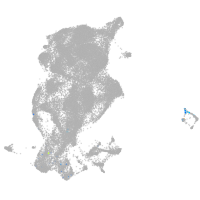 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX571955.1 | 0.141 | rnasekb | -0.032 |
| XLOC-042515 | 0.122 | atp6v0cb | -0.032 |
| BX323031.2 | 0.099 | ckbb | -0.030 |
| LOC110440051 | 0.090 | ppdpfb | -0.028 |
| CR450813.1 | 0.084 | snap25b | -0.028 |
| XLOC-028354 | 0.080 | syt5b | -0.027 |
| zgc:110216 | 0.070 | atp6v1e1b | -0.027 |
| si:ch211-113a14.24 | 0.067 | atp6v1g1 | -0.027 |
| fbxo5 | 0.064 | sypb | -0.026 |
| LOC100537297 | 0.061 | gapdhs | -0.025 |
| si:dkey-23a13.8 | 0.060 | calm1a | -0.025 |
| si:dkey-108k21.10 | 0.060 | ndrg4 | -0.024 |
| cks1b | 0.059 | vamp2 | -0.023 |
| zgc:165555.3 | 0.059 | rs1a | -0.023 |
| zgc:110425 | 0.059 | atp6ap2 | -0.023 |
| CU633933.1 | 0.057 | atpv0e2 | -0.023 |
| zgc:110540 | 0.057 | pvalb1 | -0.023 |
| mibp | 0.056 | tpi1b | -0.023 |
| ccna2 | 0.055 | rtn1a | -0.023 |
| rfc5 | 0.055 | COX3 | -0.022 |
| dut | 0.054 | mt-atp6 | -0.022 |
| zgc:153405 | 0.054 | ppp1r14c | -0.022 |
| smc2 | 0.054 | pcbp3 | -0.022 |
| rrm1 | 0.053 | zc4h2 | -0.021 |
| si:dkey-261m9.17 | 0.053 | calb2b | -0.021 |
| dek | 0.053 | pvalb2 | -0.021 |
| si:dkey-11d20.1 | 0.053 | gng3 | -0.021 |
| LO018260.2 | 0.052 | sncb | -0.021 |
| banf1 | 0.052 | atp1a3a | -0.021 |
| mki67 | 0.052 | gng13b | -0.021 |
| mier1a | 0.051 | oaz1b | -0.021 |
| chaf1a | 0.051 | mpp6b | -0.021 |
| pimr143 | 0.051 | CR383676.1 | -0.021 |
| pcna | 0.050 | mt-co2 | -0.021 |
| cd74a | 0.050 | atp1b2a | -0.020 |