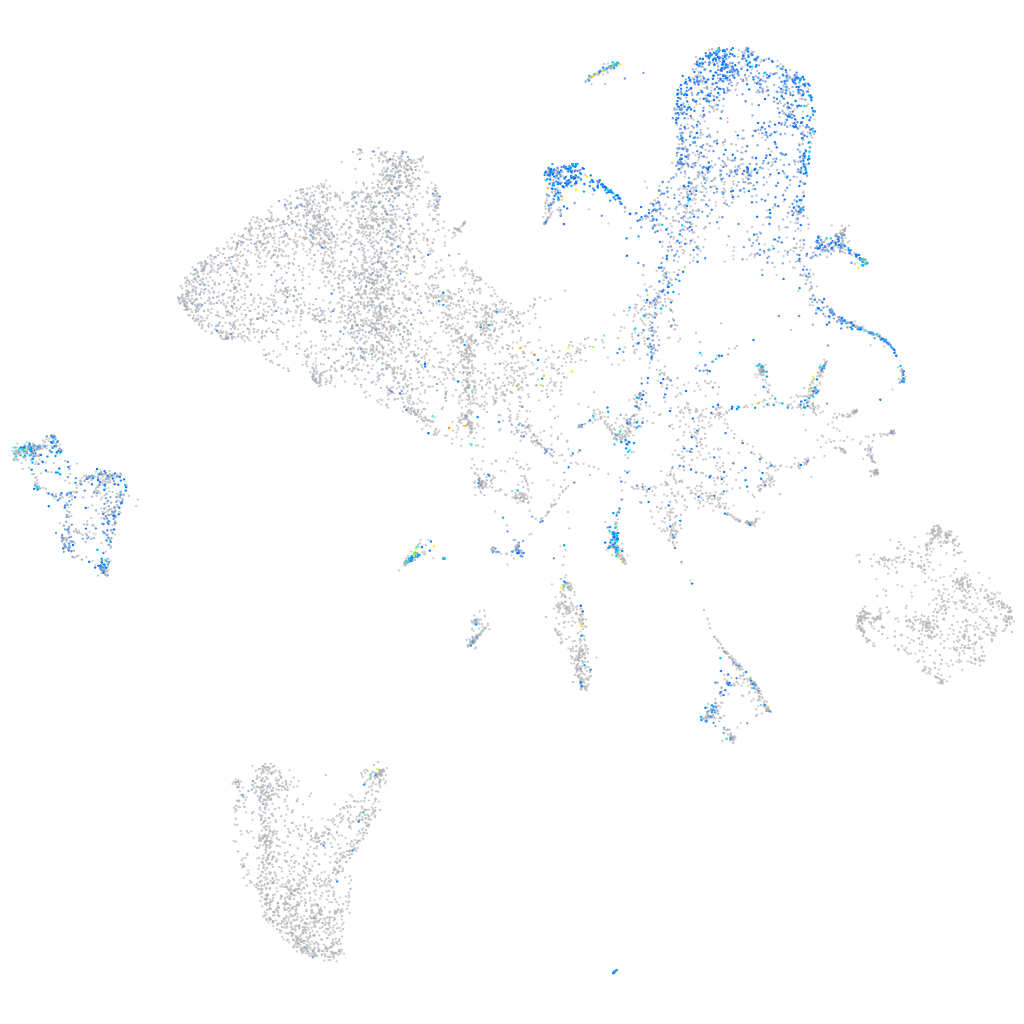
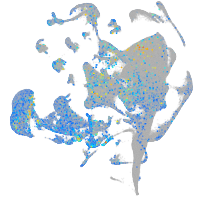
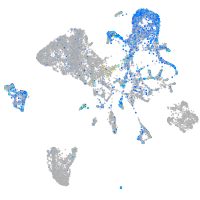
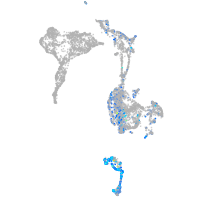
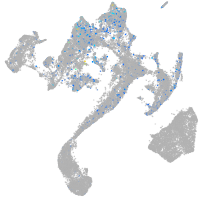
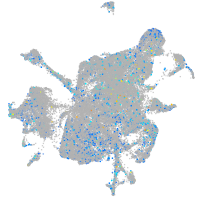
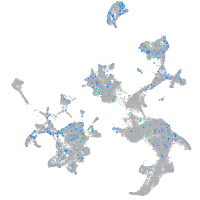
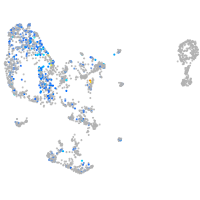
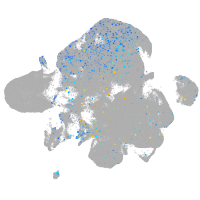
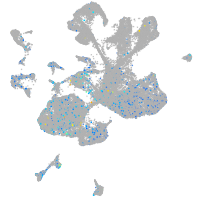
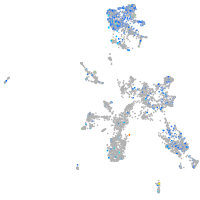
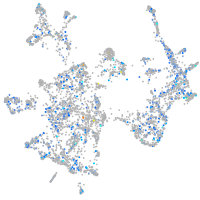
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| serpinb1l3 | 0.454 | bhmt | -0.325 |
| serpinb1 | 0.430 | aqp12 | -0.313 |
| cap1 | 0.422 | apoc1 | -0.286 |
| krt4 | 0.421 | agxtb | -0.281 |
| mvp | 0.418 | kng1 | -0.260 |
| cd63 | 0.410 | ttc36 | -0.251 |
| cldn15la | 0.410 | tfa | -0.250 |
| gsto2 | 0.405 | serpina1 | -0.249 |
| GCA | 0.401 | serpina1l | -0.247 |
| crip1 | 0.401 | apoa2 | -0.247 |
| tmsb1 | 0.398 | grhprb | -0.245 |
| eps8l3b | 0.397 | zgc:123103 | -0.244 |
| plac8.1 | 0.396 | rbp2b | -0.243 |
| si:ch211-137i24.10 | 0.395 | fabp10a | -0.241 |
| lxn | 0.394 | hpda | -0.240 |
| si:ch211-71m22.1 | 0.391 | uox | -0.240 |
| tmem176l.2 | 0.389 | cpn1 | -0.239 |
| anxa2b | 0.387 | apom | -0.238 |
| itpk1a | 0.385 | fgg | -0.238 |
| wu:fb59d01 | 0.384 | fgb | -0.236 |
| nccrp1 | 0.383 | fetub | -0.235 |
| zgc:172079 | 0.381 | hao1 | -0.235 |
| vil1 | 0.379 | ces2 | -0.235 |
| tprg1 | 0.379 | ambp | -0.234 |
| s100a10a | 0.378 | mat1a | -0.233 |
| krt92 | 0.376 | f2 | -0.232 |
| prr13 | 0.375 | fabp3 | -0.232 |
| sri | 0.375 | ttr | -0.232 |
| phlda2 | 0.374 | LOC110437731 | -0.231 |
| tmem45b | 0.372 | vtnb | -0.229 |
| si:dkeyp-73b11.8 | 0.366 | serpinf2b | -0.229 |
| wu:fb18f06 | 0.364 | c9 | -0.229 |
| si:ch1073-443f11.2 | 0.363 | gc | -0.229 |
| calml4a | 0.362 | fga | -0.228 |
| stoml3b | 0.362 | cfhl4 | -0.227 |