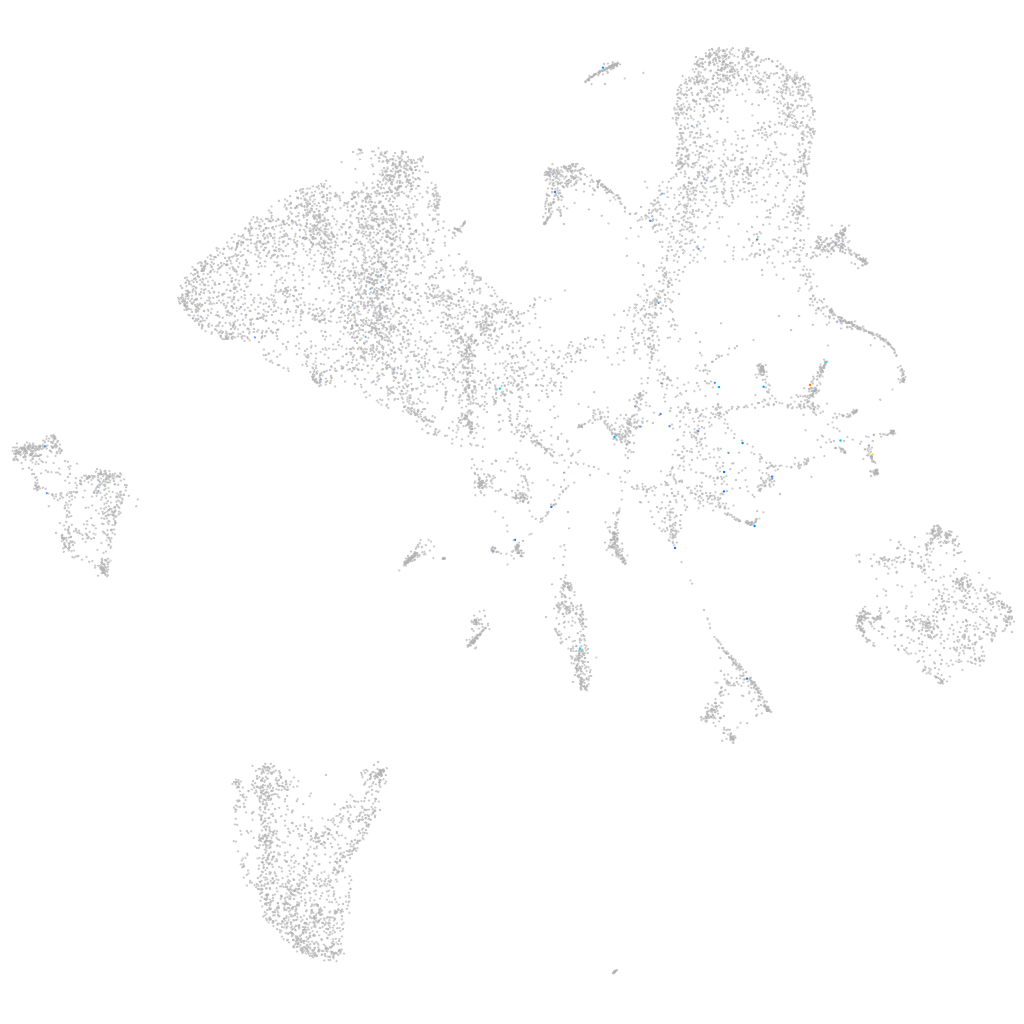
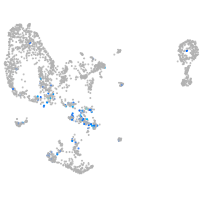
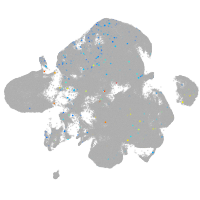
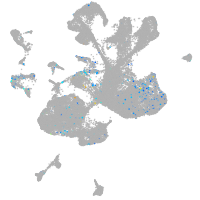
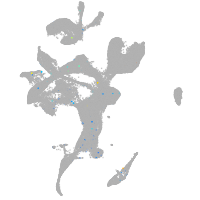
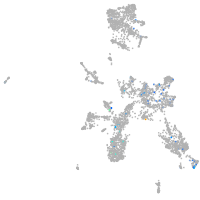
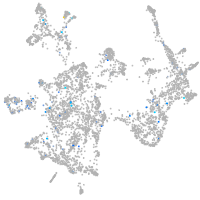
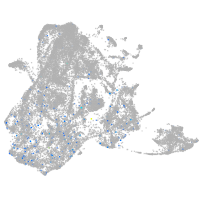
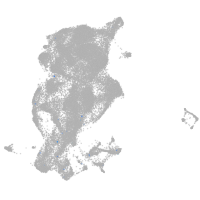
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR847897.1 | 0.295 | gamt | -0.038 |
| mitfb | 0.223 | gapdh | -0.036 |
| CR407584.1 | 0.183 | dap | -0.034 |
| gnrh2 | 0.180 | gatm | -0.034 |
| znfx1 | 0.175 | glud1b | -0.033 |
| CABZ01044764.2 | 0.171 | nupr1b | -0.032 |
| rxfp3.3a1 | 0.166 | ahcy | -0.031 |
| slc34a1a | 0.162 | pnp4b | -0.030 |
| sprn | 0.162 | pck1 | -0.030 |
| XLOC-009558 | 0.154 | eef1da | -0.029 |
| zgc:153311 | 0.154 | cox7a1 | -0.029 |
| CABZ01085139.1 | 0.153 | agxtb | -0.028 |
| lrfn4a | 0.151 | rpl6 | -0.028 |
| crybb3 | 0.151 | ucp1 | -0.028 |
| LOC100537404 | 0.150 | gpx4a | -0.027 |
| CABZ01085140.1 | 0.148 | apoa4b.1 | -0.027 |
| arhgap22 | 0.146 | abat | -0.027 |
| si:ch211-152l15.2 | 0.143 | mat1a | -0.027 |
| zgc:194990 | 0.141 | bhmt | -0.026 |
| gpr137bb | 0.137 | apoa1b | -0.026 |
| incenp | 0.136 | apoc2 | -0.026 |
| tmem245 | 0.133 | gstz1 | -0.026 |
| rab25b | 0.133 | prss1 | -0.026 |
| AL935186.2 | 0.132 | aldob | -0.026 |
| BX324164.3 | 0.131 | afp4 | -0.026 |
| BX547992.1 | 0.129 | cx32.3 | -0.026 |
| extl3 | 0.127 | faxdc2 | -0.026 |
| alkal2a | 0.126 | slc16a10 | -0.025 |
| BX927333.1 | 0.126 | eif4ebp3 | -0.025 |
| dpp6b | 0.125 | cyp2n13 | -0.025 |
| BX927234.1 | 0.122 | gda | -0.025 |
| znf385b | 0.119 | ctrb1 | -0.025 |
| LOC110440020 | 0.115 | gnmt | -0.025 |
| nalcn | 0.115 | CELA1 (1 of many) | -0.024 |
| tnfrsf18 | 0.115 | prss59.2 | -0.024 |