Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis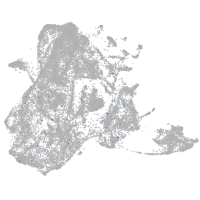 |
periderm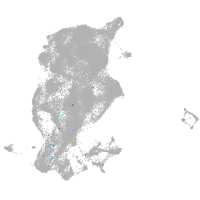 |
fin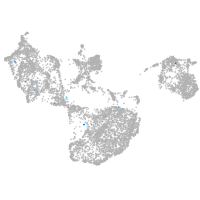 |
ionocytes / mucous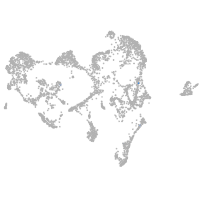 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tgm1l4 | 0.023 | ckbb | -0.016 |
| XLOC-042222 | 0.022 | elavl3 | -0.015 |
| he1.3 | 0.021 | gpm6aa | -0.015 |
| LOC103908899 | 0.021 | rtn1a | -0.015 |
| fkbp9 | 0.020 | stmn1b | -0.015 |
| pcna | 0.020 | tuba1c | -0.015 |
| selenoh | 0.020 | gpm6ab | -0.014 |
| atf3 | 0.019 | gng3 | -0.013 |
| BX927258.1 | 0.019 | pvalb1 | -0.013 |
| cdh1 | 0.019 | nova2 | -0.012 |
| chaf1a | 0.019 | pvalb2 | -0.012 |
| krt92 | 0.019 | tmsb | -0.012 |
| nr2f5 | 0.019 | tuba1a | -0.012 |
| XLOC-032526 | 0.019 | atp6v0cb | -0.011 |
| zfp36l1a | 0.019 | fez1 | -0.011 |
| asph | 0.018 | myt1b | -0.011 |
| CABZ01075068.1 | 0.018 | rnasekb | -0.011 |
| dut | 0.018 | rtn1b | -0.011 |
| junba | 0.018 | atp6v1e1b | -0.010 |
| lig1 | 0.018 | CU467822.1 | -0.010 |
| mcm4 | 0.018 | elavl4 | -0.010 |
| mcm6 | 0.018 | myt1a | -0.010 |
| rpa1 | 0.018 | scrt2 | -0.010 |
| rpa2 | 0.018 | si:dkeyp-75h12.5 | -0.010 |
| sdc4 | 0.018 | sncb | -0.010 |
| XLOC-039121 | 0.018 | vamp2 | -0.010 |
| cdca7b | 0.017 | CU634008.1 | -0.010 |
| fen1 | 0.017 | dpysl2b | -0.010 |
| hdlbpa | 0.017 | ccni | -0.009 |
| id1 | 0.017 | celf2 | -0.009 |
| mcm7 | 0.017 | celf3a | -0.009 |
| naalad2 | 0.017 | csdc2a | -0.009 |
| orc4 | 0.017 | dpysl3 | -0.009 |
| rpa3 | 0.017 | fyna | -0.009 |
| si:dkey-195m11.8 | 0.017 | gap43 | -0.009 |




