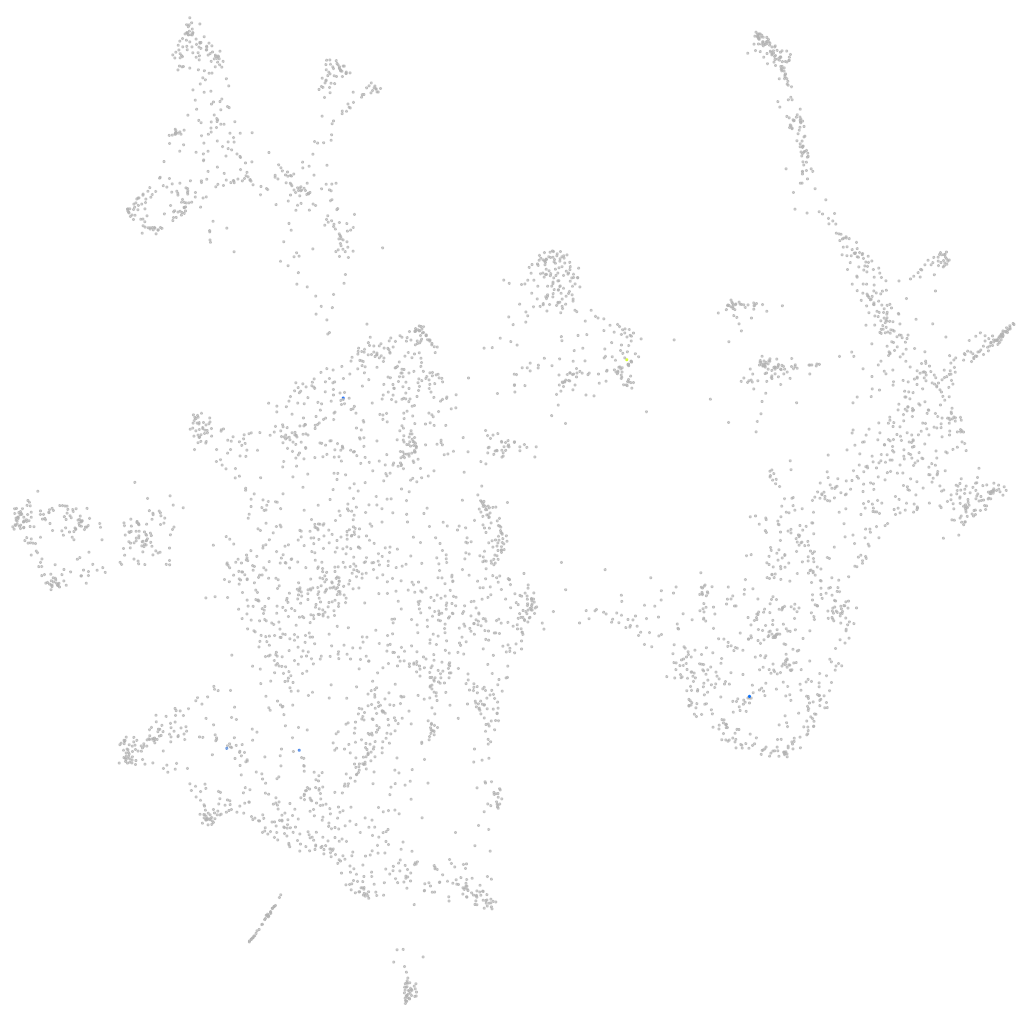
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mep1a.2 | 0.866 | sec61g | -0.036 |
| cx32.2 | 0.685 | NC-002333.4 | -0.031 |
| slc18a3b | 0.678 | mt-nd4 | -0.029 |
| LOC101885014 | 0.522 | ndufb9 | -0.027 |
| pln | 0.508 | ap2m1a | -0.026 |
| rgs2 | 0.424 | lsm6 | -0.026 |
| apnl | 0.416 | fkbp1aa | -0.025 |
| cfbl | 0.381 | psmb6 | -0.025 |
| il4 | 0.378 | ssr3 | -0.025 |
| nmt1b | 0.358 | selenof | -0.024 |
| abcb11b | 0.334 | ndufb4 | -0.024 |
| slc47a1 | 0.331 | serf2 | -0.023 |
| si:ch211-212d10.1 | 0.330 | psmb1 | -0.023 |
| CR751227.1 | 0.322 | uqcrfs1 | -0.023 |
| sb:cb37 | 0.321 | arhgdia | -0.023 |
| tm6sf2 | 0.292 | drap1 | -0.022 |
| wscd1b | 0.286 | oaz1a | -0.022 |
| ces3 | 0.284 | cct2 | -0.022 |
| LOC101887100 | 0.282 | eif3ha | -0.022 |
| acp5b | 0.281 | capzb | -0.021 |
| pla2g1b | 0.281 | rbx1 | -0.021 |
| six6a | 0.281 | rab2a | -0.021 |
| slc15a1b | 0.280 | cct3 | -0.021 |
| comtd1 | 0.253 | pdia3 | -0.021 |
| tirap | 0.244 | gng5 | -0.020 |
| zgc:112265 | 0.244 | arf2b | -0.020 |
| AL845362.1 | 0.242 | eya1 | -0.020 |
| gabrg1 | 0.240 | mgst3a | -0.020 |
| si:ch73-306e8.2 | 0.235 | calm1a | -0.020 |
| XLOC-017089 | 0.226 | ndufa3 | -0.020 |
| col6a4a | 0.225 | tbca | -0.020 |
| si:ch211-270n8.1 | 0.222 | zgc:56676 | -0.020 |
| XLOC-019150 | 0.217 | snrpf | -0.020 |
| slco1c1 | 0.215 | sub1a | -0.020 |
| RF00056 | 0.212 | ndufa10 | -0.020 |