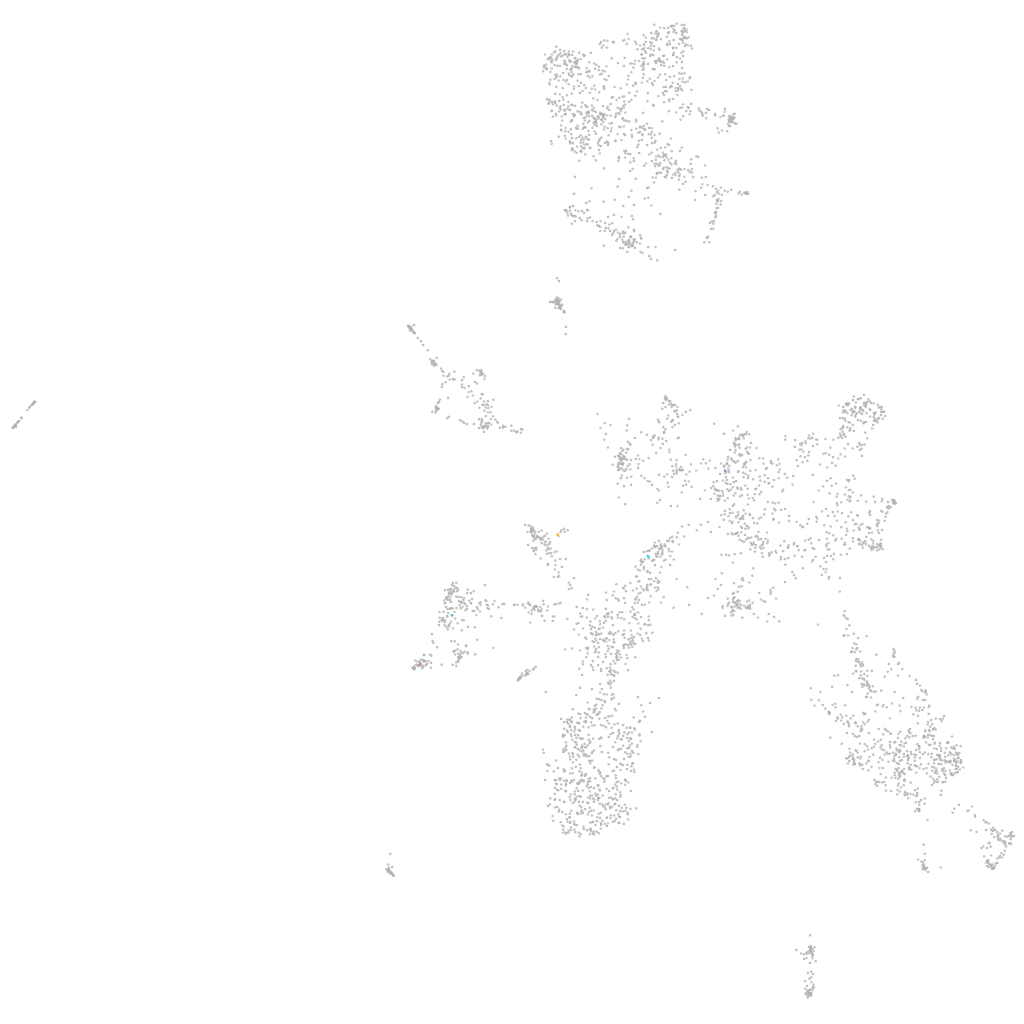
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| actn2b | 0.664 | slc25a5 | -0.063 |
| itgb3b | 0.505 | rps2 | -0.050 |
| znf1128 | 0.458 | eif3f | -0.046 |
| nek8 | 0.457 | rsl24d1 | -0.045 |
| eogt | 0.446 | edf1 | -0.043 |
| prelp | 0.442 | ubb | -0.043 |
| colgalt2 | 0.405 | atp5f1b | -0.042 |
| grip2b | 0.364 | epcam | -0.040 |
| irx1b | 0.343 | rpl9 | -0.040 |
| fam155a | 0.336 | anxa11b | -0.039 |
| rasl11a | 0.336 | atp5f1c | -0.039 |
| creb5b | 0.336 | ube2v1 | -0.038 |
| twist2 | 0.334 | arhgdia | -0.038 |
| XLOC-029603 | 0.332 | rbx1 | -0.038 |
| egfl7 | 0.329 | eef1db | -0.037 |
| mrc2 | 0.322 | actb1 | -0.037 |
| cplx3b | 0.314 | rpl5b | -0.037 |
| gtpbp3 | 0.301 | rack1 | -0.036 |
| clcn5b | 0.300 | btf3 | -0.036 |
| rhcgb | 0.289 | ndufb8 | -0.036 |
| pth1rb | 0.282 | mcl1a | -0.036 |
| gdpd3b | 0.274 | myl12.1 | -0.035 |
| kctd16a | 0.269 | cct6a | -0.035 |
| cyp2k21 | 0.261 | cyc1 | -0.035 |
| XLOC-013065 | 0.261 | dbi | -0.035 |
| CU972512.1 | 0.243 | taf12 | -0.035 |
| RIC3 | 0.238 | mycbp | -0.034 |
| st3gal5 | 0.237 | ccni | -0.034 |
| nefma | 0.235 | xbp1 | -0.034 |
| ogna | 0.222 | arf2b | -0.034 |
| nebl | 0.210 | ddx5 | -0.034 |
| LOC101882843 | 0.203 | mibp2 | -0.034 |
| ism1 | 0.203 | syf2 | -0.034 |
| loxl1 | 0.199 | hnrnpabb | -0.034 |
| tnfsf10l | 0.198 | NC-002333.17 | -0.034 |