Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 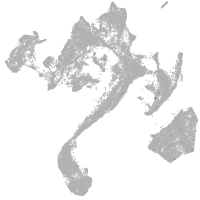 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 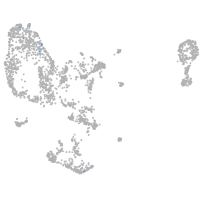 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis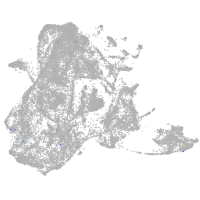 |
periderm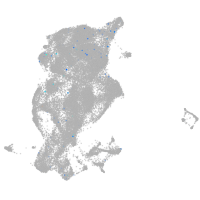 |
fin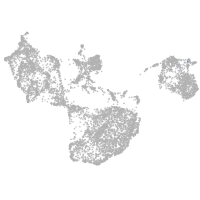 |
ionocytes / mucous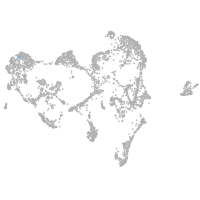 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aldob | 0.037 | tuba1c | -0.022 |
| dhrs13a.2 | 0.029 | marcksl1a | -0.022 |
| hhla2b.2 | 0.029 | gpm6aa | -0.020 |
| mgst1.2 | 0.029 | nova2 | -0.019 |
| si:ch73-52f15.5 | 0.029 | rtn1a | -0.019 |
| tmem45b | 0.029 | ckbb | -0.018 |
| cmah | 0.028 | elavl3 | -0.018 |
| krt17 | 0.028 | gpm6ab | -0.018 |
| mid1ip1a | 0.028 | stmn1b | -0.018 |
| scel | 0.028 | fabp3 | -0.017 |
| si:dkey-33i11.9 | 0.028 | fam168a | -0.017 |
| myo15b | 0.027 | rnasekb | -0.017 |
| zgc:111983 | 0.027 | atp6v0cb | -0.016 |
| anxa1b | 0.026 | gapdhs | -0.016 |
| cd9b | 0.026 | hbbe1.3 | -0.016 |
| cldne | 0.026 | hmgb3a | -0.016 |
| eps8l3a | 0.026 | jpt1b | -0.016 |
| IZUMO1R | 0.026 | mdkb | -0.016 |
| lye | 0.026 | gng3 | -0.015 |
| mgst2 | 0.026 | hbae3 | -0.015 |
| nqo1 | 0.026 | sncb | -0.015 |
| ponzr2 | 0.026 | tuba1a | -0.015 |
| prr13 | 0.026 | dpysl2b | -0.015 |
| rnf183 | 0.026 | celf2 | -0.014 |
| si:ch211-157c3.4 | 0.026 | cspg5a | -0.014 |
| si:ch211-207n23.2 | 0.026 | elavl4 | -0.014 |
| si:ch211-79k12.1 | 0.026 | gnao1a | -0.014 |
| si:ch73-204p21.2 | 0.026 | hbae1.1 | -0.014 |
| si:dkey-247k7.2 | 0.026 | hmgb1b | -0.014 |
| stard14 | 0.026 | si:dkeyp-75h12.5 | -0.014 |
| stx11b.1 | 0.026 | tmeff1b | -0.014 |
| bin2a | 0.025 | tmsb | -0.014 |
| chs1 | 0.025 | tubb5 | -0.014 |
| CR855996.2 | 0.025 | vamp2 | -0.014 |
| gsta.1 | 0.025 | wu:fb55g09 | -0.014 |




