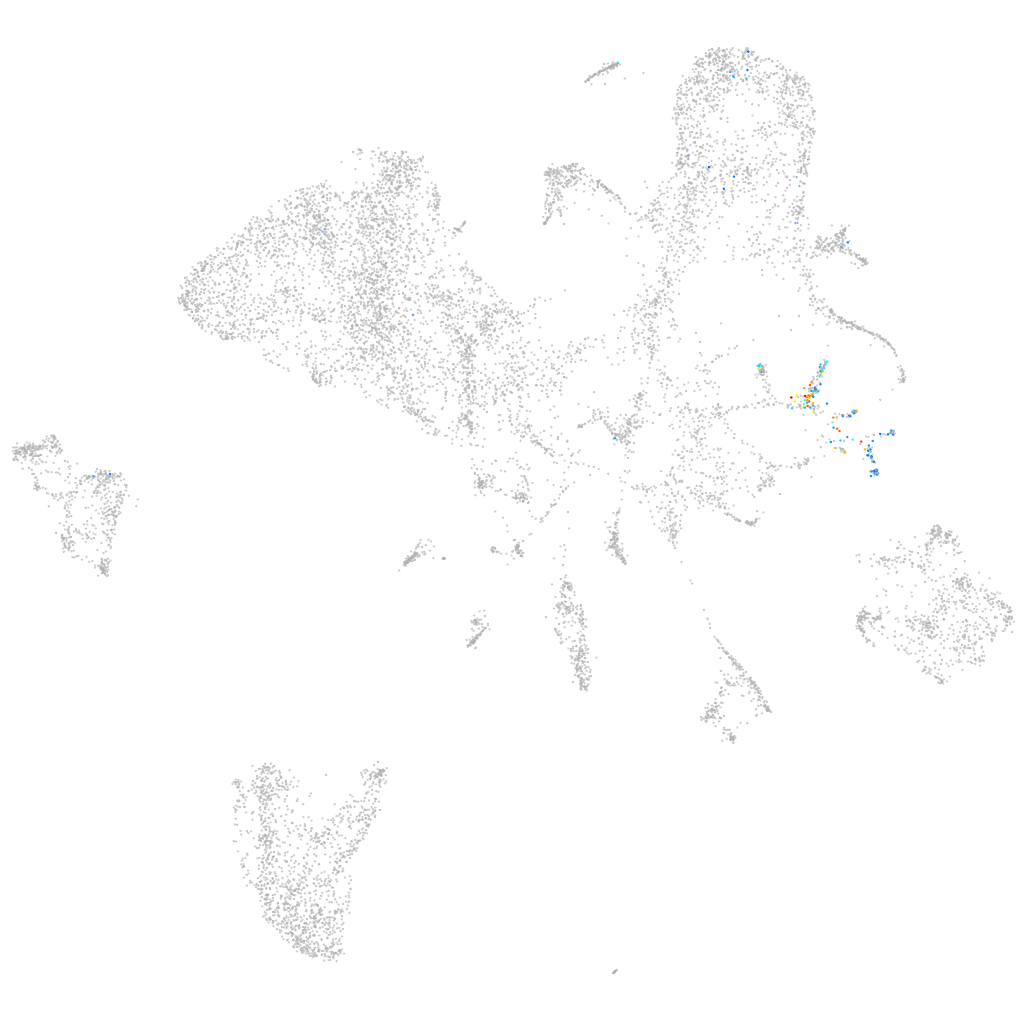
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| arxa | 0.462 | ahcy | -0.112 |
| rprmb | 0.436 | gapdh | -0.100 |
| CR556712.1 | 0.422 | fabp3 | -0.093 |
| calca | 0.410 | hdlbpa | -0.092 |
| scgn | 0.404 | gamt | -0.089 |
| kcnj11 | 0.387 | bhmt | -0.086 |
| neurod1 | 0.351 | gatm | -0.082 |
| scg3 | 0.345 | aldh6a1 | -0.081 |
| syt1a | 0.332 | eif4ebp1 | -0.079 |
| rem2 | 0.328 | mat1a | -0.079 |
| c2cd4a | 0.328 | fbp1b | -0.077 |
| si:dkey-153k10.9 | 0.326 | si:dkey-16p21.8 | -0.077 |
| isl1 | 0.322 | aldob | -0.076 |
| pcsk1nl | 0.315 | rpl4 | -0.076 |
| rasd4 | 0.310 | glud1b | -0.076 |
| mir7a-1 | 0.306 | gstr | -0.075 |
| trmt9b | 0.304 | rps20 | -0.075 |
| vat1 | 0.299 | apoa1b | -0.074 |
| pax6b | 0.295 | apoa4b.1 | -0.074 |
| insl5a | 0.294 | apoc1 | -0.073 |
| insm1a | 0.293 | aqp12 | -0.073 |
| tspan7b | 0.290 | cx32.3 | -0.072 |
| mir375-2 | 0.288 | rpl6 | -0.072 |
| oprd1b | 0.288 | gstp1 | -0.072 |
| abcc8 | 0.278 | gstt1a | -0.072 |
| kcnk9 | 0.277 | aldh7a1 | -0.071 |
| capsla | 0.277 | adka | -0.071 |
| nkx2.2a | 0.276 | apoc2 | -0.071 |
| gpc1a | 0.270 | eno3 | -0.071 |
| pcsk2 | 0.264 | agxta | -0.071 |
| scg2b | 0.262 | agxtb | -0.070 |
| mlnl | 0.261 | cebpd | -0.069 |
| etv1 | 0.260 | shmt1 | -0.068 |
| egr4 | 0.259 | wu:fj16a03 | -0.068 |
| slc35g2b | 0.257 | gnmt | -0.068 |