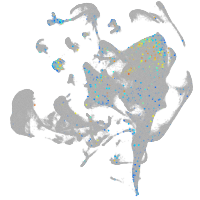
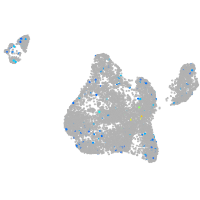
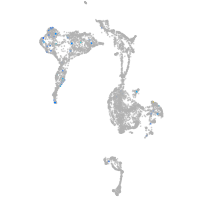
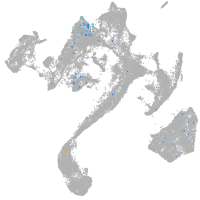
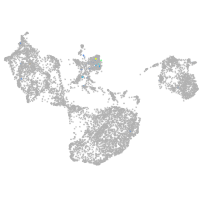
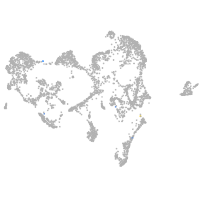
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01078244.2 | 0.164 | CABZ01021592.1 | -0.113 |
| si:ch73-265h17.4 | 0.155 | si:dkey-251i10.2 | -0.098 |
| zmynd10 | 0.153 | uraha | -0.093 |
| pax1a | 0.150 | paics | -0.083 |
| CU074419.1 | 0.145 | tmem130 | -0.081 |
| plekhg6 | 0.142 | slc22a7a | -0.073 |
| CU459012.1 | 0.139 | si:ch211-251b21.1 | -0.072 |
| gabrr2b | 0.135 | aox5 | -0.071 |
| kcnk9 | 0.133 | gch2 | -0.071 |
| rxfp3 | 0.133 | cyb5a | -0.066 |
| BX465864.1 | 0.131 | bco1 | -0.066 |
| LOC103911288 | 0.128 | bscl2l | -0.062 |
| znfl1g | 0.126 | sult1st1 | -0.062 |
| si:dkey-37g12.1 | 0.122 | mdh1aa | -0.061 |
| ulk4 | 0.121 | sprb | -0.059 |
| agrp | 0.120 | aqp3a | -0.059 |
| spra | 0.116 | rbp4l | -0.058 |
| plxnc1 | 0.116 | aldob | -0.058 |
| gaa3-as | 0.115 | prdx5 | -0.058 |
| CABZ01088864.1 | 0.114 | prdx1 | -0.056 |
| tyrp1a | 0.113 | prtfdc1 | -0.056 |
| oca2 | 0.112 | pax7b | -0.056 |
| shank3b | 0.112 | mibp | -0.055 |
| dct | 0.112 | cax1 | -0.054 |
| XLOC-026582 | 0.111 | zgc:113337 | -0.053 |
| shroom3 | 0.111 | sigmar1 | -0.053 |
| anxa2a | 0.110 | TMEM19 | -0.052 |
| pmela | 0.110 | slc2a11b | -0.051 |
| si:ch211-197n1.2 | 0.110 | cx30.3 | -0.050 |
| caprin2 | 0.110 | slc2a15a | -0.050 |
| grxcr1a.1 | 0.110 | si:zfos-1069f5.1 | -0.049 |
| tyrp1b | 0.109 | aldh1l1 | -0.049 |
| phf19 | 0.109 | pts | -0.049 |
| gig2j | 0.109 | asip2b | -0.048 |
| XLOC-011085 | 0.109 | oacyl | -0.048 |