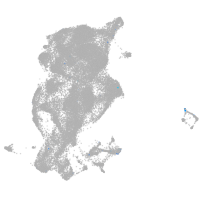
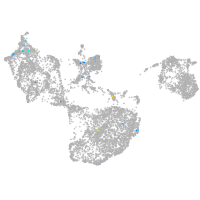
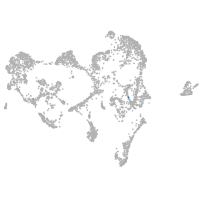
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gcgb | 0.291 | ahcy | -0.061 |
| pnoca | 0.270 | gapdh | -0.054 |
| scgn | 0.262 | gamt | -0.053 |
| dkk3b | 0.254 | aldob | -0.051 |
| cngb3.1 | 0.247 | gatm | -0.050 |
| rims4 | 0.245 | eno3 | -0.048 |
| kcnj11 | 0.244 | mat1a | -0.047 |
| si:dkey-47k16.4 | 0.243 | aldh7a1 | -0.046 |
| LOC100537384 | 0.240 | glud1b | -0.045 |
| elfn1a | 0.240 | fbp1b | -0.045 |
| pcdh1a3 | 0.237 | aqp12 | -0.044 |
| ak5l | 0.229 | bhmt | -0.044 |
| arxa | 0.223 | hdlbpa | -0.043 |
| CR774179.5 | 0.221 | aldh6a1 | -0.042 |
| gcga | 0.220 | CABZ01032488.1 | -0.042 |
| rprmb | 0.217 | apoc2 | -0.042 |
| pax6b | 0.213 | gstr | -0.042 |
| si:dkey-153k10.9 | 0.213 | igf2bp3 | -0.041 |
| neurod1 | 0.211 | si:dkey-16p21.8 | -0.041 |
| LOC103911631 | 0.209 | cx32.3 | -0.041 |
| syt1a | 0.209 | gstt1a | -0.041 |
| rasd4 | 0.208 | shmt1 | -0.041 |
| cacna1c | 0.204 | tktb | -0.041 |
| LOC108180103 | 0.203 | krt8 | -0.041 |
| CU929403.2 | 0.202 | cebpb | -0.040 |
| cntnap2a | 0.201 | afp4 | -0.040 |
| pcdh2ab1 | 0.199 | lipf | -0.040 |
| rims2a | 0.198 | cebpd | -0.040 |
| tspan7b | 0.194 | hspd1 | -0.039 |
| isl1 | 0.190 | scp2a | -0.039 |
| cadm2a | 0.190 | gstp1 | -0.039 |
| fam49bb | 0.189 | sec61a1 | -0.039 |
| fam69ab | 0.185 | g6pca.2 | -0.039 |
| slc30a2 | 0.184 | ppa1b | -0.039 |
| npb | 0.184 | slc16a10 | -0.039 |