Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm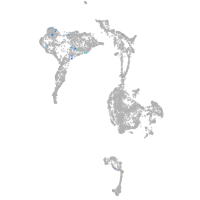 |
muscle 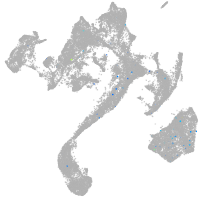 |
mural cells / non-skeletal muscle 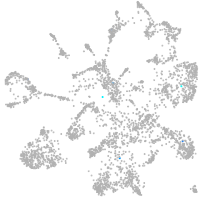 |
mesenchyme 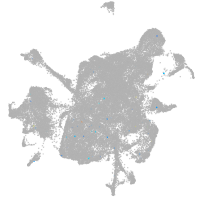 |
hematopoietic / vasculature 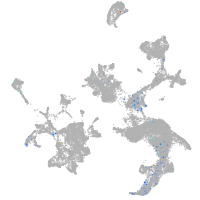 |
pronephros 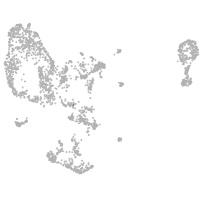 |
neural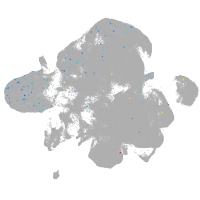 |
spinal cord / glia |
eye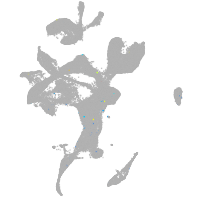 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU695223.1 | 0.767 | sf3b2 | -0.219 |
| pnp5b | 0.765 | eif5a | -0.213 |
| efr3ba | 0.748 | nme2b.1 | -0.212 |
| emilin2a | 0.748 | CABZ01044053.1 | -0.186 |
| kif3cb | 0.748 | hnrnpa1a | -0.183 |
| si:ch1073-513e17.1 | 0.748 | AK6 | -0.168 |
| si:dkey-1h24.2 | 0.748 | ddx18 | -0.166 |
| disp1 | 0.709 | psmb4 | -0.163 |
| cenpv | 0.680 | tomm22 | -0.163 |
| lifra | 0.673 | rnf20 | -0.162 |
| ugt1b5 | 0.672 | nudt21 | -0.158 |
| CABZ01085594.1 | 0.654 | srrt | -0.157 |
| FP085398.1 | 0.654 | gar1 | -0.155 |
| sept4b | 0.643 | hnrnpd | -0.154 |
| LOC108181079 | 0.641 | dvl3a | -0.154 |
| hoxa13b | 0.634 | tlk1a | -0.153 |
| LOC108181166 | 0.627 | fxr1 | -0.153 |
| kita | 0.611 | mphosph8 | -0.153 |
| CYTL1 | 0.610 | nap1l1 | -0.151 |
| CT025771.1 | 0.608 | rrp36 | -0.149 |
| exoc3l1 | 0.601 | rpf1 | -0.148 |
| znf692 | 0.591 | tuba8l | -0.148 |
| RNF122 | 0.588 | mkrn1 | -0.148 |
| epn3a | 0.582 | snrpa1 | -0.147 |
| gal3st4 | 0.582 | ssrp1a | -0.147 |
| fam171b | 0.580 | ncaph | -0.147 |
| SH3RF3 | 0.573 | clptm1 | -0.147 |
| pdss2 | 0.570 | eif5 | -0.146 |
| tgfbr3 | 0.566 | pelp1 | -0.145 |
| pdlim4 | 0.565 | srpk1a | -0.145 |
| eef2k | 0.562 | slc25a5 | -0.144 |
| filip1l | 0.562 | sec62 | -0.144 |
| arhgap36 | 0.561 | cct4 | -0.144 |
| samd4a | 0.558 | twf1a | -0.143 |
| si:ch73-89b15.3 | 0.557 | thop1 | -0.143 |