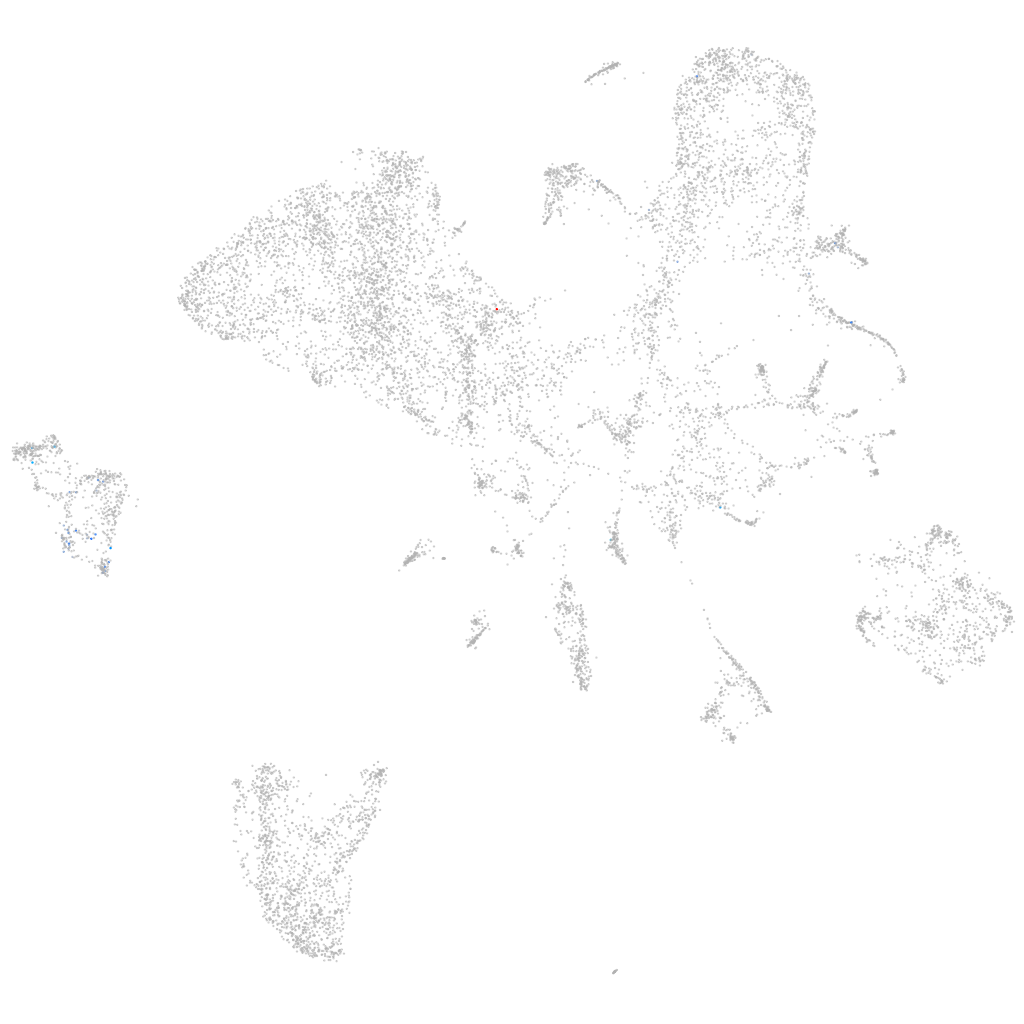
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-044324 | 0.501 | gatm | -0.038 |
| CABZ01068367.1 | 0.496 | mat1a | -0.034 |
| slc2a3b | 0.409 | hspe1 | -0.034 |
| si:dkey-5n7.2 | 0.359 | rpl23 | -0.033 |
| gpr37a | 0.352 | pklr | -0.032 |
| BX649411.2 | 0.331 | pcbd1 | -0.032 |
| sox10 | 0.326 | gpd1b | -0.031 |
| mbpa | 0.299 | scp2a | -0.031 |
| zwi | 0.271 | agxta | -0.031 |
| cd59 | 0.269 | abat | -0.030 |
| pacs2 | 0.266 | cx32.3 | -0.030 |
| CABZ01092746.1 | 0.245 | kng1 | -0.029 |
| ZBTB26 | 0.242 | gcshb | -0.029 |
| gldn | 0.236 | pnp4b | -0.029 |
| nrgna | 0.227 | bhmt | -0.029 |
| gas2l3 | 0.221 | gnmt | -0.028 |
| BX323854.2 | 0.218 | vdac3 | -0.028 |
| CT027609.1 | 0.213 | tfa | -0.028 |
| LO017718.1 | 0.212 | rltgr | -0.028 |
| plp1a | 0.208 | fth1a | -0.027 |
| cldnk | 0.196 | ftcd | -0.027 |
| crygm2d15 | 0.192 | nipsnap3a | -0.027 |
| AL935186.9.1 | 0.192 | sult2st2 | -0.027 |
| AL845295.1 | 0.188 | slco1d1 | -0.027 |
| zgc:113278 | 0.185 | ttc36 | -0.027 |
| dennd4b | 0.177 | msrb2 | -0.027 |
| pigl | 0.177 | si:dkey-16p21.8 | -0.027 |
| cysltr3 | 0.176 | serpina1l | -0.027 |
| scube1 | 0.168 | serpinf2b | -0.027 |
| gria2a | 0.165 | vtnb | -0.027 |
| BX537104.1 | 0.165 | fgg | -0.026 |
| si:ch73-119p20.1 | 0.163 | phyhd1 | -0.026 |
| zgc:172053 | 0.162 | cdo1 | -0.026 |
| amn1 | 0.159 | tdo2a | -0.026 |
| ecrg4a | 0.152 | ambp | -0.026 |