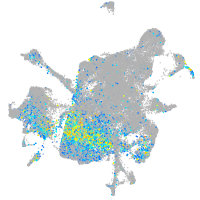
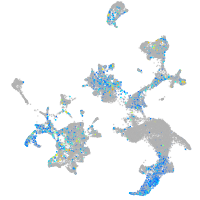
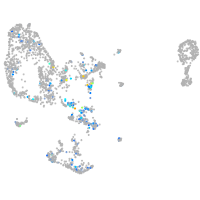
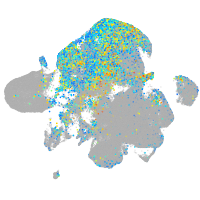
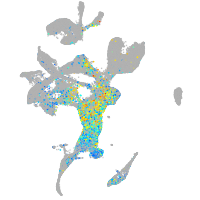
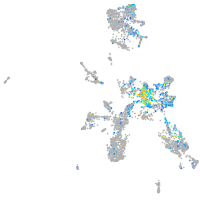
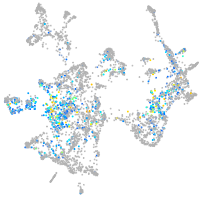
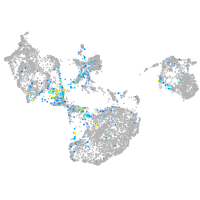
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110540 | 0.530 | txn | -0.264 |
| dut | 0.504 | gstp1 | -0.255 |
| mibp | 0.504 | gapdhs | -0.255 |
| CABZ01005379.1 | 0.493 | anxa5b | -0.243 |
| rrm1 | 0.486 | tmem59 | -0.225 |
| slbp | 0.486 | selenow2b | -0.215 |
| stmn1a | 0.484 | selenow1 | -0.211 |
| fbxo5 | 0.481 | calm1a | -0.209 |
| cks1b | 0.480 | bik | -0.207 |
| esco2 | 0.478 | nfe2l2a | -0.202 |
| nasp | 0.463 | fxyd1 | -0.200 |
| pcna | 0.459 | cldnh | -0.198 |
| chaf1a | 0.447 | atp1b1a | -0.198 |
| lbr | 0.441 | wu:fb18f06 | -0.198 |
| zgc:110216 | 0.439 | smdt1b | -0.197 |
| ccna2 | 0.436 | itm2ba | -0.194 |
| si:dkey-261m9.17 | 0.432 | CR383676.1 | -0.191 |
| rpa2 | 0.430 | ccni | -0.189 |
| haus4 | 0.428 | gabarapa | -0.188 |
| tk1 | 0.422 | rnasekb | -0.186 |
| si:dkey-6i22.5 | 0.421 | vamp2 | -0.186 |
| dek | 0.421 | atp6v1e1b | -0.185 |
| cdca5 | 0.420 | chga | -0.185 |
| mis12 | 0.419 | mt-co2 | -0.182 |
| mki67 | 0.409 | pvalb5 | -0.180 |
| rpa3 | 0.407 | msi2b | -0.179 |
| hist1h4l | 0.407 | si:ch211-153b23.5 | -0.176 |
| lig1 | 0.406 | gnb1a | -0.176 |
| zgc:165555.3 | 0.402 | zgc:165573 | -0.174 |
| mcm7 | 0.401 | gsto2 | -0.174 |
| smc2 | 0.400 | nupr1a | -0.173 |
| her2 | 0.399 | b2ml | -0.173 |
| her15.1 | 0.394 | htatip2 | -0.171 |
| asf1ba | 0.391 | si:ch1073-443f11.2 | -0.171 |
| XLOC-042899 | 0.389 | zgc:65894 | -0.171 |