KRR1 small subunit processome component homolog
ZFIN 
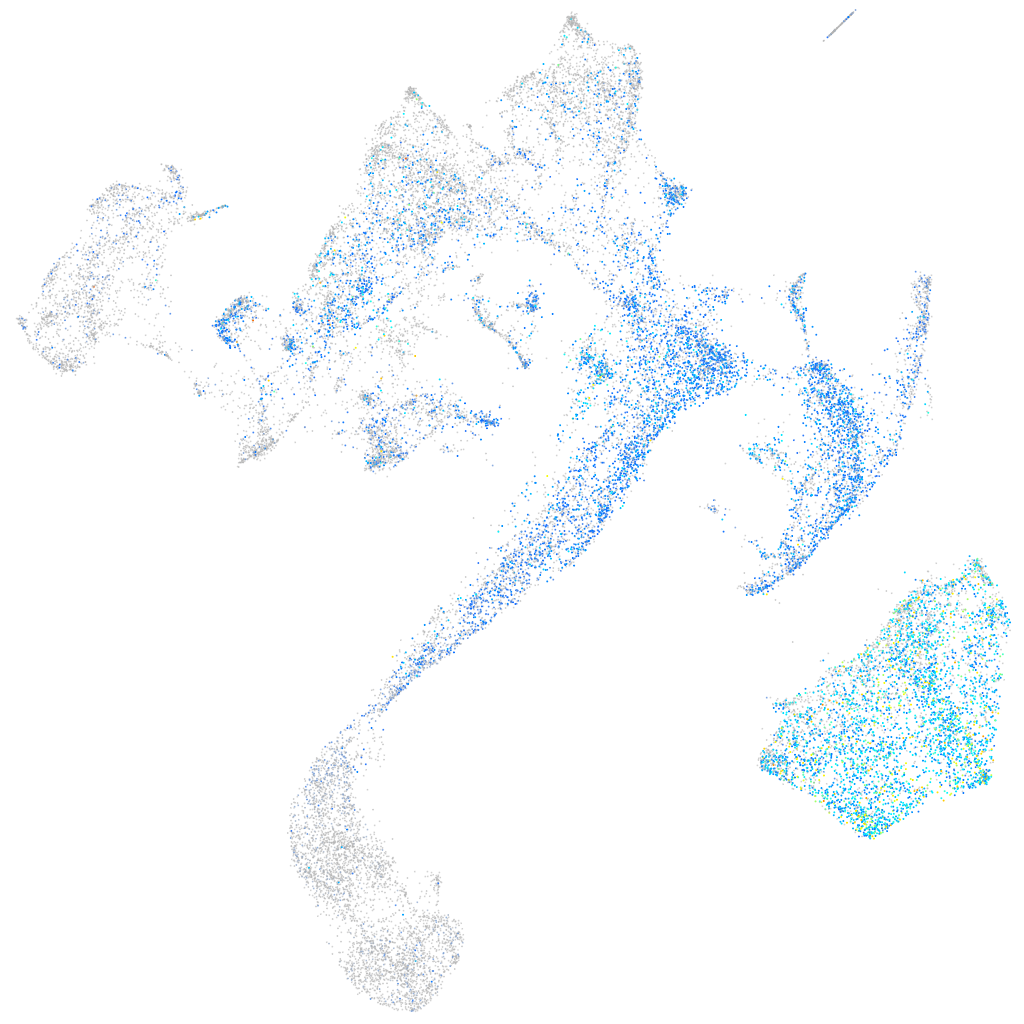
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ncl | 0.488 | rpl37 | -0.413 |
| nop58 | 0.483 | zgc:114188 | -0.382 |
| fbl | 0.477 | rps10 | -0.375 |
| dkc1 | 0.472 | rps17 | -0.358 |
| anp32e | 0.471 | actc1b | -0.333 |
| npm1a | 0.470 | fabp3 | -0.304 |
| hnrnpa1b | 0.470 | mt-nd1 | -0.302 |
| nop56 | 0.464 | ckma | -0.298 |
| hnrnpub | 0.462 | ak1 | -0.298 |
| hnrnpabb | 0.456 | ckmb | -0.296 |
| ppig | 0.452 | bhmt | -0.294 |
| stm | 0.450 | zgc:158463 | -0.292 |
| rbm4.3 | 0.445 | atp2a1 | -0.289 |
| ilf3b | 0.442 | vdac3 | -0.289 |
| syncrip | 0.441 | idh2 | -0.285 |
| srsf1a | 0.441 | mt-atp6 | -0.283 |
| nop2 | 0.437 | tnnc2 | -0.283 |
| top1l | 0.434 | ttn.2 | -0.282 |
| ebna1bp2 | 0.430 | neb | -0.280 |
| nucks1a | 0.430 | pvalb1 | -0.278 |
| hnrnpa0b | 0.429 | aldoab | -0.278 |
| seta | 0.429 | eno3 | -0.277 |
| hmga1a | 0.428 | atp5meb | -0.276 |
| safb | 0.427 | mylpfa | -0.276 |
| marcksb | 0.427 | eef1da | -0.276 |
| brd3a | 0.427 | tpi1b | -0.275 |
| ddx18 | 0.426 | pvalb2 | -0.271 |
| bms1 | 0.426 | nme2b.2 | -0.270 |
| snrpa | 0.424 | acta1b | -0.267 |
| gar1 | 0.424 | eif4a1b | -0.266 |
| snrnp70 | 0.423 | actn3a | -0.266 |
| hnrnpa1a | 0.421 | actn3b | -0.265 |
| khdrbs1a | 0.420 | ldb3b | -0.265 |
| anp32b | 0.418 | COX3 | -0.265 |
| sf3b2 | 0.417 | si:ch73-367p23.2 | -0.265 |