lysine methyltransferase 5Ab
ZFIN 
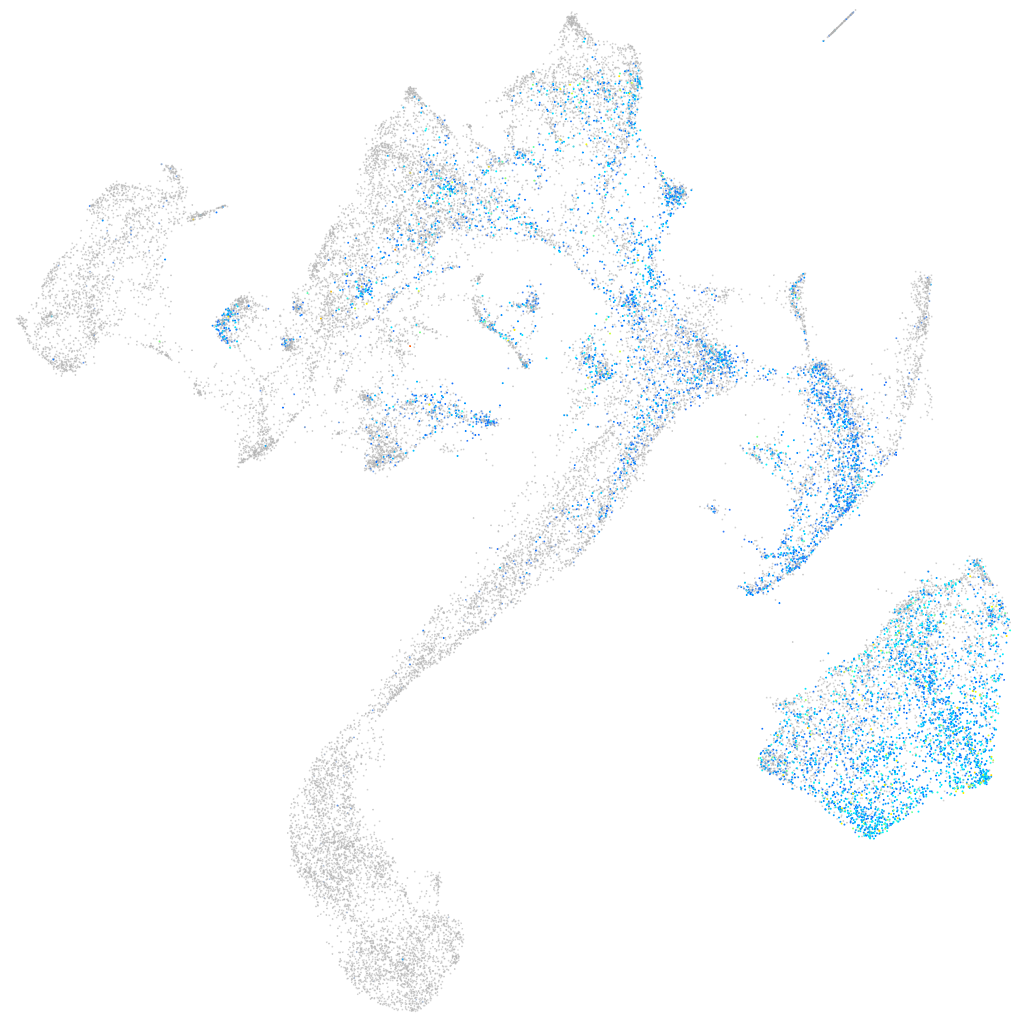
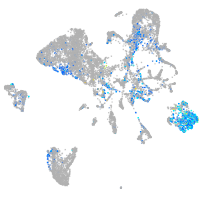
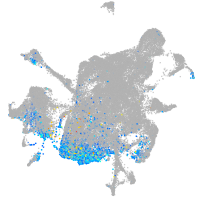
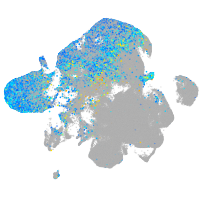
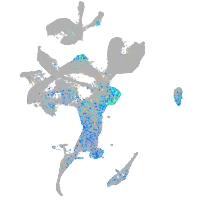
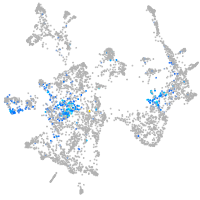
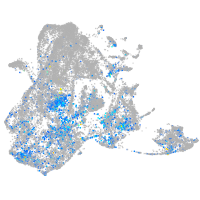
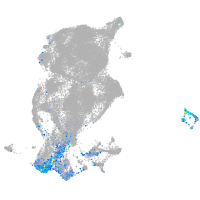
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ube2c | 0.636 | rpl37 | -0.355 |
| ccnb1 | 0.629 | zgc:114188 | -0.337 |
| tpx2 | 0.616 | actc1b | -0.332 |
| si:ch211-69g19.2 | 0.588 | rps10 | -0.328 |
| cdca8 | 0.581 | rps17 | -0.316 |
| mki67 | 0.577 | mt-nd1 | -0.297 |
| plk1 | 0.570 | ttn.2 | -0.294 |
| nusap1 | 0.567 | ttn.1 | -0.285 |
| mad2l1 | 0.562 | vdac3 | -0.285 |
| cdc20 | 0.550 | ak1 | -0.282 |
| kif11 | 0.541 | aldoab | -0.280 |
| kpna2 | 0.539 | atp5if1b | -0.274 |
| cdk1 | 0.535 | fabp3 | -0.274 |
| cenpf | 0.519 | ckmb | -0.273 |
| top2a | 0.518 | atp2a1 | -0.273 |
| birc5a | 0.518 | ckma | -0.273 |
| dlgap5 | 0.508 | tmem38a | -0.272 |
| aurka | 0.505 | mt-atp6 | -0.269 |
| kifc1 | 0.502 | atp5meb | -0.269 |
| aurkb | 0.502 | pabpc4 | -0.267 |
| aspm | 0.501 | tnnc2 | -0.266 |
| ccna2 | 0.499 | suclg1 | -0.261 |
| ccnb3 | 0.486 | acta1b | -0.260 |
| kif23 | 0.481 | eef1da | -0.260 |
| prc1a | 0.481 | desma | -0.260 |
| hmgb2a | 0.464 | mylpfa | -0.259 |
| top1l | 0.456 | atp5f1b | -0.259 |
| anp32a | 0.448 | tpma | -0.258 |
| cdc14b | 0.445 | acta1a | -0.257 |
| g2e3 | 0.439 | atp5mc3b | -0.257 |
| kif22 | 0.438 | gamt | -0.256 |
| fam76b | 0.433 | mybphb | -0.256 |
| numa1 | 0.432 | eno1a | -0.255 |
| seta | 0.429 | neb | -0.254 |
| hnrnpa1b | 0.429 | srl | -0.251 |