kinesin family member 3Cb
ZFIN 
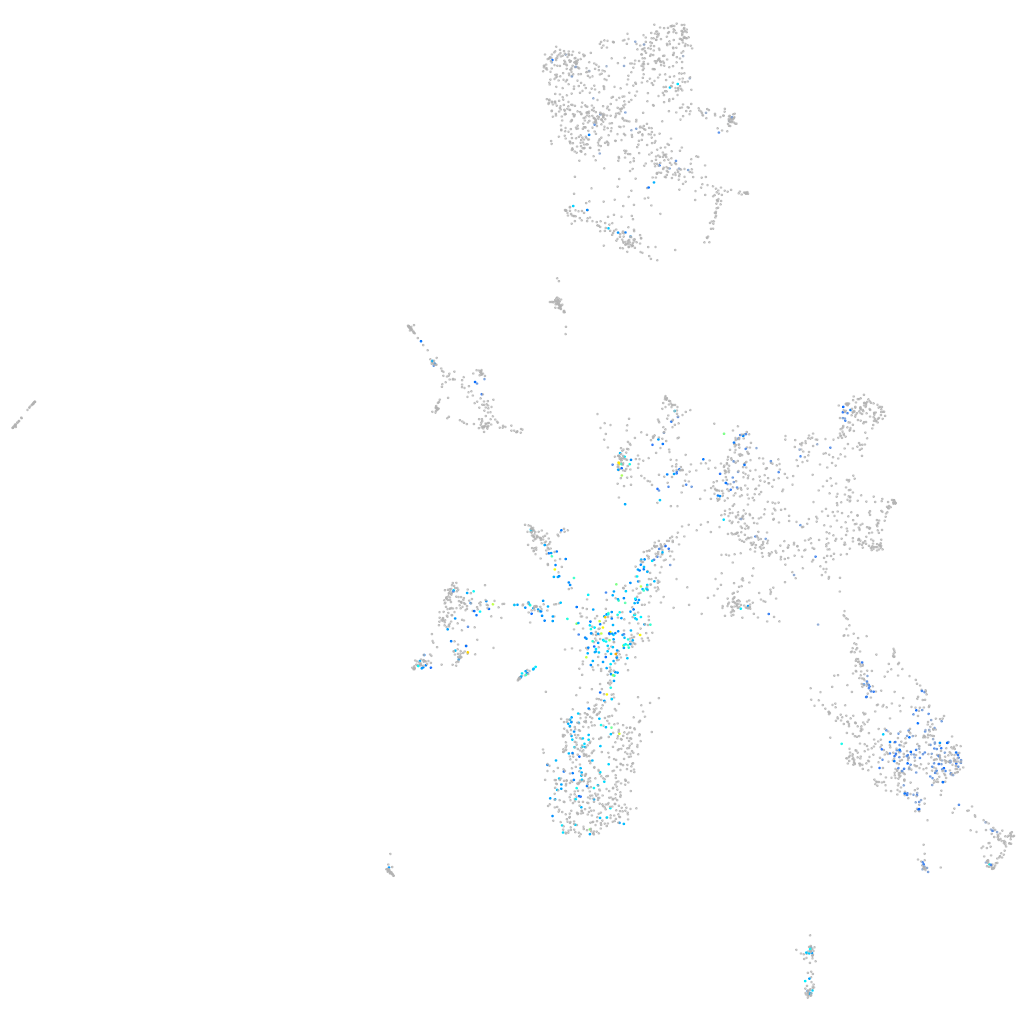
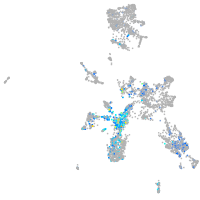
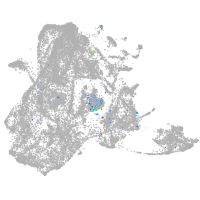
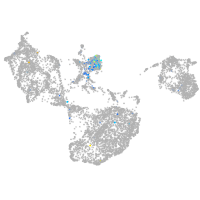
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stmn1b | 0.398 | krt8 | -0.213 |
| si:dkeyp-75h12.5 | 0.340 | fosab | -0.206 |
| LOC100537342 | 0.337 | krt18a.1 | -0.201 |
| elavl4 | 0.335 | pfn1 | -0.197 |
| cd99l2 | 0.331 | eef1da | -0.191 |
| tmsb | 0.328 | si:ch211-286b5.5 | -0.187 |
| rtn1a | 0.326 | sdc4 | -0.187 |
| tmeff1b | 0.323 | btg2 | -0.183 |
| ppp1r14ba | 0.321 | s100v2 | -0.180 |
| zgc:65894 | 0.321 | sox3 | -0.180 |
| gng3 | 0.320 | lgals3b | -0.180 |
| tspan2a | 0.312 | ahnak | -0.178 |
| si:dkey-276j7.1 | 0.305 | COX7A2 (1 of many) | -0.176 |
| elavl3 | 0.304 | zgc:158343 | -0.174 |
| dpysl2b | 0.297 | ier2b | -0.173 |
| zc4h2 | 0.294 | sparc | -0.172 |
| gap43 | 0.294 | plecb | -0.171 |
| insm1a | 0.294 | sox2 | -0.171 |
| klf7a | 0.290 | tagln2 | -0.170 |
| dpysl3 | 0.289 | krt4 | -0.170 |
| stmn2a | 0.285 | GCA | -0.164 |
| sncb | 0.284 | her9 | -0.163 |
| maptb | 0.281 | cfl1l | -0.163 |
| map7d2b | 0.280 | s100u | -0.162 |
| tuba1c | 0.274 | cldni | -0.162 |
| ebf3a | 0.273 | egr1 | -0.162 |
| cnrip1a | 0.273 | anxa4 | -0.161 |
| epb41a | 0.270 | s100a10b | -0.160 |
| vdac3 | 0.269 | fgfbp1b | -0.160 |
| gsna | 0.268 | rpz5 | -0.160 |
| si:ch211-10a23.2 | 0.266 | apoeb | -0.160 |
| tuba1a | 0.263 | junba | -0.160 |
| atf5a | 0.263 | arhgef5 | -0.160 |
| csdc2a | 0.263 | s100v1 | -0.158 |
| gdi1 | 0.262 | krt97 | -0.157 |