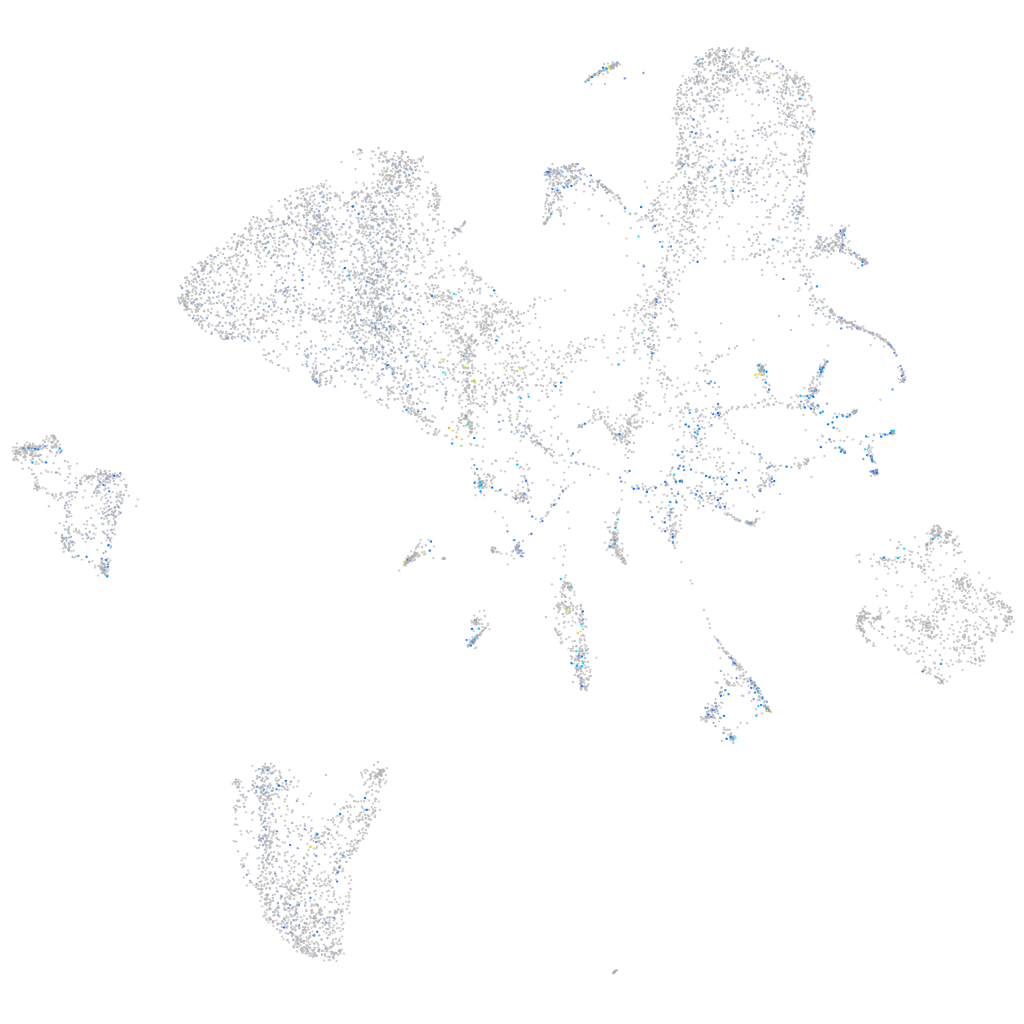
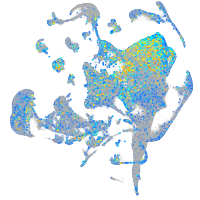
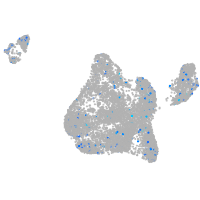
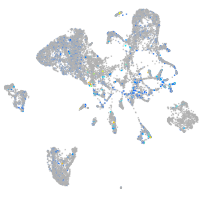
kinesin family member 1B
ZFIN 
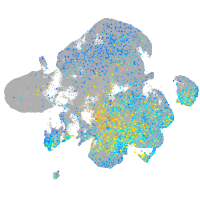
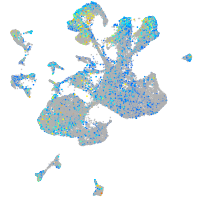
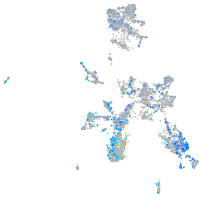
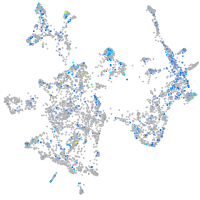
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc35g2b | 0.156 | gapdh | -0.091 |
| scg3 | 0.155 | ahcy | -0.084 |
| aldocb | 0.155 | gamt | -0.077 |
| cpe | 0.148 | gatm | -0.066 |
| scg2b | 0.147 | aldob | -0.066 |
| cxxc5a | 0.145 | gstt1a | -0.065 |
| scgn | 0.144 | scp2a | -0.064 |
| vat1 | 0.143 | mat1a | -0.064 |
| gapdhs | 0.142 | gcshb | -0.063 |
| syt1a | 0.140 | fbp1b | -0.063 |
| rnasekb | 0.139 | sult2st2 | -0.061 |
| pcsk1 | 0.139 | pnp4b | -0.059 |
| lysmd2 | 0.136 | glud1b | -0.057 |
| cnrip1a | 0.136 | agxtb | -0.057 |
| mir7a-1 | 0.133 | apoc1 | -0.056 |
| atp6ap1a | 0.132 | bhmt | -0.056 |
| scg2a | 0.131 | apoc2 | -0.056 |
| pax6b | 0.131 | nipsnap3a | -0.056 |
| tspan7b | 0.129 | dap | -0.055 |
| tmem145 | 0.128 | cox7a1 | -0.055 |
| jpt1b | 0.128 | suclg1 | -0.055 |
| vamp2 | 0.128 | ucp1 | -0.054 |
| neurod1 | 0.128 | rdh1 | -0.054 |
| cplx2l | 0.127 | afp4 | -0.054 |
| zgc:92818 | 0.126 | apoa4b.1 | -0.053 |
| pcsk1nl | 0.124 | etnppl | -0.053 |
| gabrz | 0.124 | sult3st2 | -0.052 |
| nrsn1 | 0.124 | gar1 | -0.052 |
| CR383676.1 | 0.124 | sod2 | -0.052 |
| cnih2 | 0.123 | cox8b | -0.052 |
| LOC108179280 | 0.122 | vox | -0.051 |
| LOC110438234 | 0.122 | acmsd | -0.051 |
| si:dkey-153k10.9 | 0.122 | eci2 | -0.051 |
| kcnj11 | 0.122 | fabp10a | -0.051 |
| unc13ba | 0.121 | gstr | -0.051 |