kinesin family member 13Ba
ZFIN 
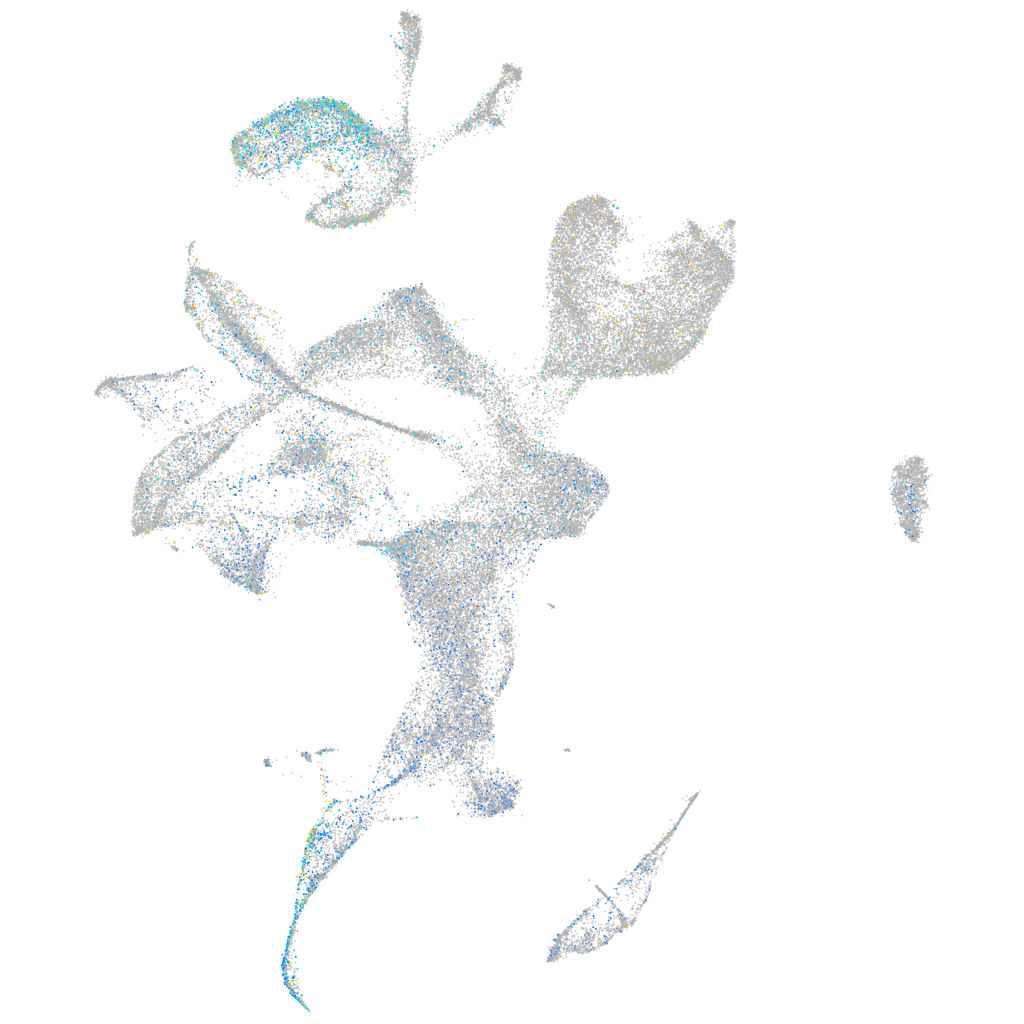
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmelb | 0.170 | hmgb2a | -0.096 |
| tspan10 | 0.169 | anp32e | -0.075 |
| tyrp1b | 0.168 | syt5b | -0.073 |
| oca2 | 0.165 | si:ch73-1a9.3 | -0.070 |
| pmela | 0.164 | neurod4 | -0.069 |
| rab27a | 0.163 | crx | -0.068 |
| dct | 0.163 | histh1l | -0.064 |
| bace2 | 0.163 | ptmab | -0.060 |
| pax10 | 0.159 | vsx1 | -0.060 |
| tyrp1a | 0.159 | rs1a | -0.057 |
| sytl2a | 0.156 | epb41a | -0.054 |
| cracr2ab | 0.155 | gnb3a | -0.052 |
| rab38 | 0.153 | samsn1a | -0.052 |
| gpr143 | 0.150 | pcna | -0.051 |
| tspan36 | 0.149 | rrm1 | -0.051 |
| LOC103910009 | 0.148 | si:ch73-281n10.2 | -0.050 |
| tyr | 0.147 | ndrg1b | -0.049 |
| col4a5 | 0.145 | stmn1a | -0.049 |
| rgrb | 0.145 | nrn1lb | -0.049 |
| slc45a2 | 0.145 | nasp | -0.048 |
| slc24a5 | 0.143 | chaf1a | -0.048 |
| msnb | 0.143 | dek | -0.047 |
| slc7a2 | 0.143 | slbp | -0.047 |
| fabp11b | 0.142 | ppp1r1b | -0.046 |
| selenou1b | 0.142 | gng13b | -0.046 |
| pttg1ipb | 0.140 | dut | -0.046 |
| tfr1a | 0.140 | CABZ01005379.1 | -0.046 |
| slc41a1 | 0.138 | rrm2 | -0.046 |
| tmem98 | 0.137 | fabp7a | -0.046 |
| tm6sf2 | 0.136 | neurod1 | -0.045 |
| meis2b | 0.135 | fen1 | -0.045 |
| zgc:110789 | 0.135 | hmgn2 | -0.045 |
| hps1 | 0.135 | vamp1 | -0.044 |
| pah | 0.134 | pcp4l1 | -0.044 |
| col4a6 | 0.134 | si:ch1073-83n3.2 | -0.044 |