KDEL endoplasmic reticulum protein retention receptor 2a
ZFIN 
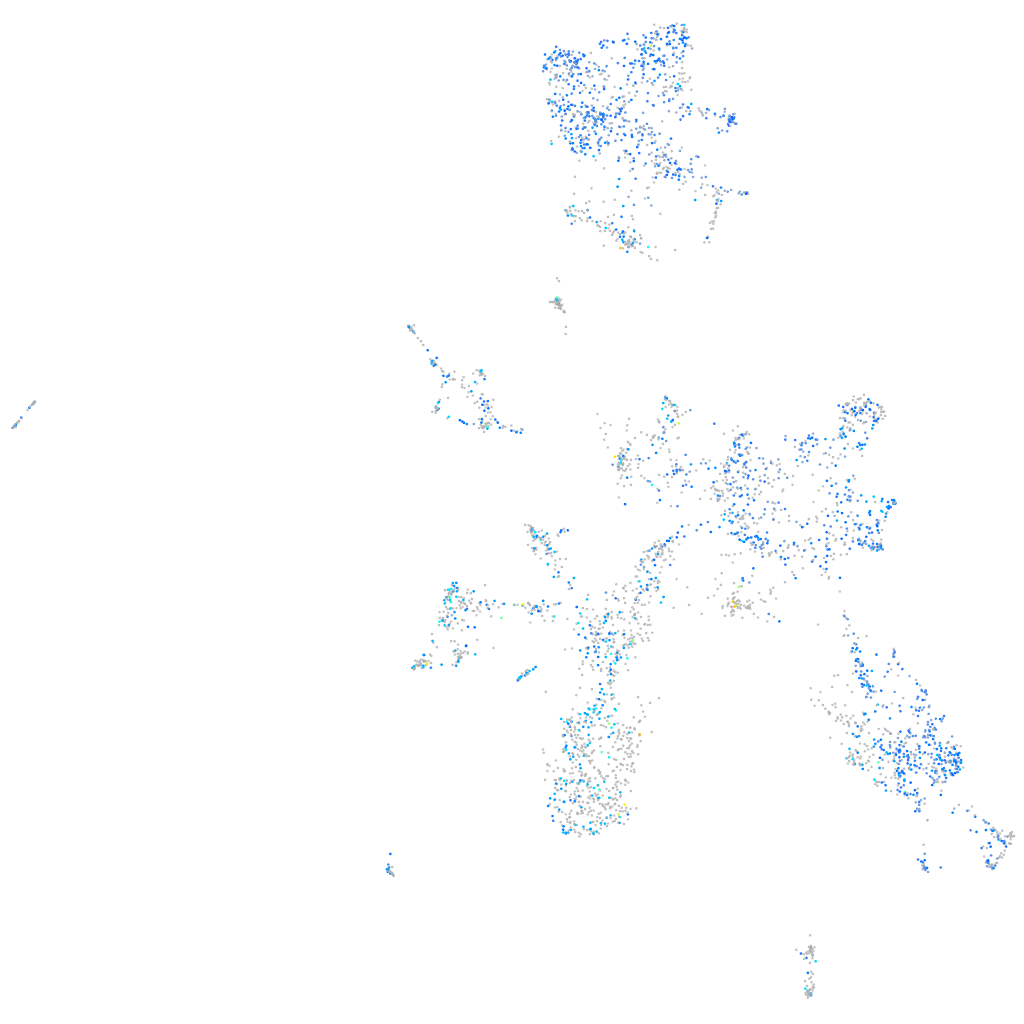
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ssr3 | 0.159 | hbbe1.1 | -0.079 |
| dad1 | 0.154 | hbbe2 | -0.071 |
| xbp1 | 0.151 | hbbe1.3 | -0.065 |
| cd9b | 0.151 | NC-002333.4 | -0.063 |
| arf2b | 0.149 | tnnt3b | -0.059 |
| copb2 | 0.149 | LOC110439372 | -0.056 |
| zgc:153867 | 0.148 | syt17 | -0.054 |
| cavin2b | 0.144 | tcnba | -0.054 |
| zgc:158343 | 0.144 | rpl38 | -0.053 |
| tubb4b | 0.144 | agbl2 | -0.052 |
| cxcl14 | 0.140 | actc1b | -0.051 |
| si:ch211-286b5.5 | 0.140 | vsnl1a | -0.050 |
| fgfbp1b | 0.139 | NC-002333.17 | -0.050 |
| flot2a | 0.139 | nrn1a | -0.050 |
| cd63 | 0.139 | stxbp5l | -0.049 |
| selenof | 0.138 | pbx3b | -0.048 |
| tmed2 | 0.138 | hbae3 | -0.047 |
| abracl | 0.136 | vopp1 | -0.047 |
| prss60.2 | 0.135 | zgc:158463 | -0.046 |
| smox | 0.135 | pvalb1 | -0.045 |
| lasp1 | 0.134 | cldn17 | -0.045 |
| zgc:165423 | 0.134 | CT583728.23 | -0.045 |
| myl12.1 | 0.134 | cox4i2 | -0.045 |
| rpz5 | 0.134 | CR925761.4 | -0.045 |
| ssr4 | 0.133 | dab1a | -0.045 |
| cst14a.2 | 0.133 | CABZ01101984.1 | -0.045 |
| cldn7b | 0.133 | lyz | -0.045 |
| s100v1 | 0.133 | mir124-3 | -0.044 |
| her9 | 0.132 | dpydb | -0.044 |
| agr1 | 0.132 | dnah12 | -0.043 |
| krtcap2 | 0.132 | lrrc74b | -0.043 |
| cdc42l | 0.132 | kif9 | -0.043 |
| arpc3 | 0.131 | LOC101886078 | -0.043 |
| ssr2 | 0.129 | CABZ01021592.1 | -0.043 |
| zgc:101744 | 0.129 | spag1b | -0.043 |