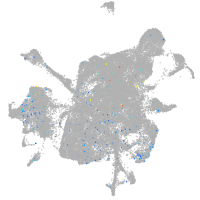
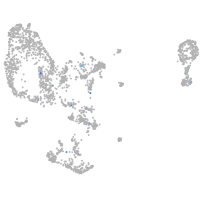
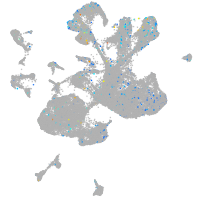
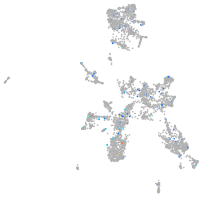
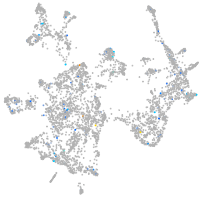
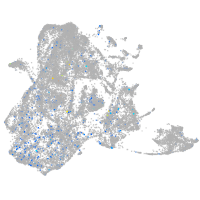
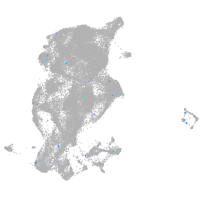
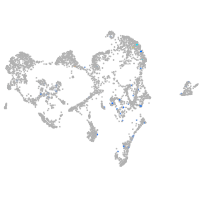
potassium channel tetramerization domain containing 6a
ZFIN 
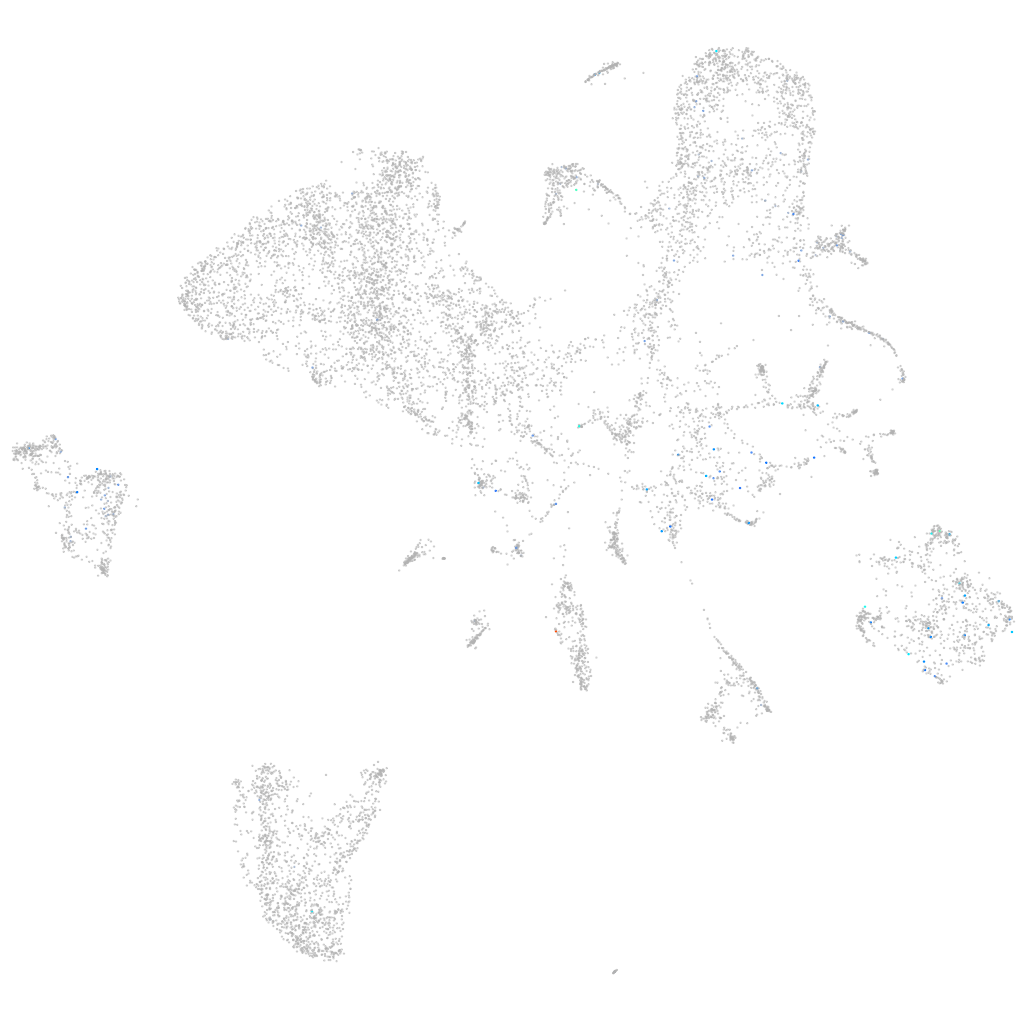
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mettl11b | 0.275 | ahcy | -0.075 |
| crygm2d20 | 0.255 | gamt | -0.067 |
| enox1 | 0.187 | rps10 | -0.063 |
| mst1rb | 0.186 | sod1 | -0.062 |
| ntng1a | 0.179 | bhmt | -0.061 |
| si:ch211-76m11.7 | 0.173 | gapdh | -0.060 |
| LOC110438299 | 0.164 | mat1a | -0.060 |
| gpr143 | 0.158 | atp5if1b | -0.059 |
| alas2 | 0.156 | zgc:92744 | -0.059 |
| BX072576.3 | 0.156 | rps17 | -0.059 |
| BX322603.1 | 0.156 | nupr1b | -0.057 |
| mrvi1 | 0.149 | apoa1b | -0.057 |
| LOC103910826 | 0.149 | rpl37 | -0.057 |
| kntc1 | 0.144 | agxtb | -0.056 |
| CABZ01057496.1 | 0.143 | suclg1 | -0.056 |
| ly6m2 | 0.140 | dap | -0.056 |
| LOC110439873 | 0.139 | apoa4b.1 | -0.056 |
| ttc9b | 0.139 | mdh1aa | -0.055 |
| ugt1b3 | 0.138 | cx32.3 | -0.055 |
| neto1 | 0.132 | gatm | -0.055 |
| BX901962.4 | 0.131 | agxta | -0.055 |
| CR377211.4 | 0.131 | prdx2 | -0.055 |
| XLOC-014770 | 0.130 | grhprb | -0.055 |
| dpysl5b | 0.130 | abat | -0.054 |
| prlh | 0.125 | gpx4a | -0.054 |
| si:ch211-113e8.5 | 0.123 | eno3 | -0.054 |
| CR626884.5 | 0.121 | ndrg2 | -0.054 |
| cryba1l1 | 0.117 | apoa2 | -0.054 |
| CABZ01078767.1 | 0.116 | scp2a | -0.053 |
| LOC100334422 | 0.115 | gstt1a | -0.053 |
| si:dkey-28d5.12 | 0.114 | aldob | -0.052 |
| scrt2 | 0.113 | ftcd | -0.052 |
| LOC110438490 | 0.111 | aldh7a1 | -0.052 |
| BX293991.1 | 0.109 | gnmt | -0.052 |
| CABZ01063203.1 | 0.108 | rbp2b | -0.052 |