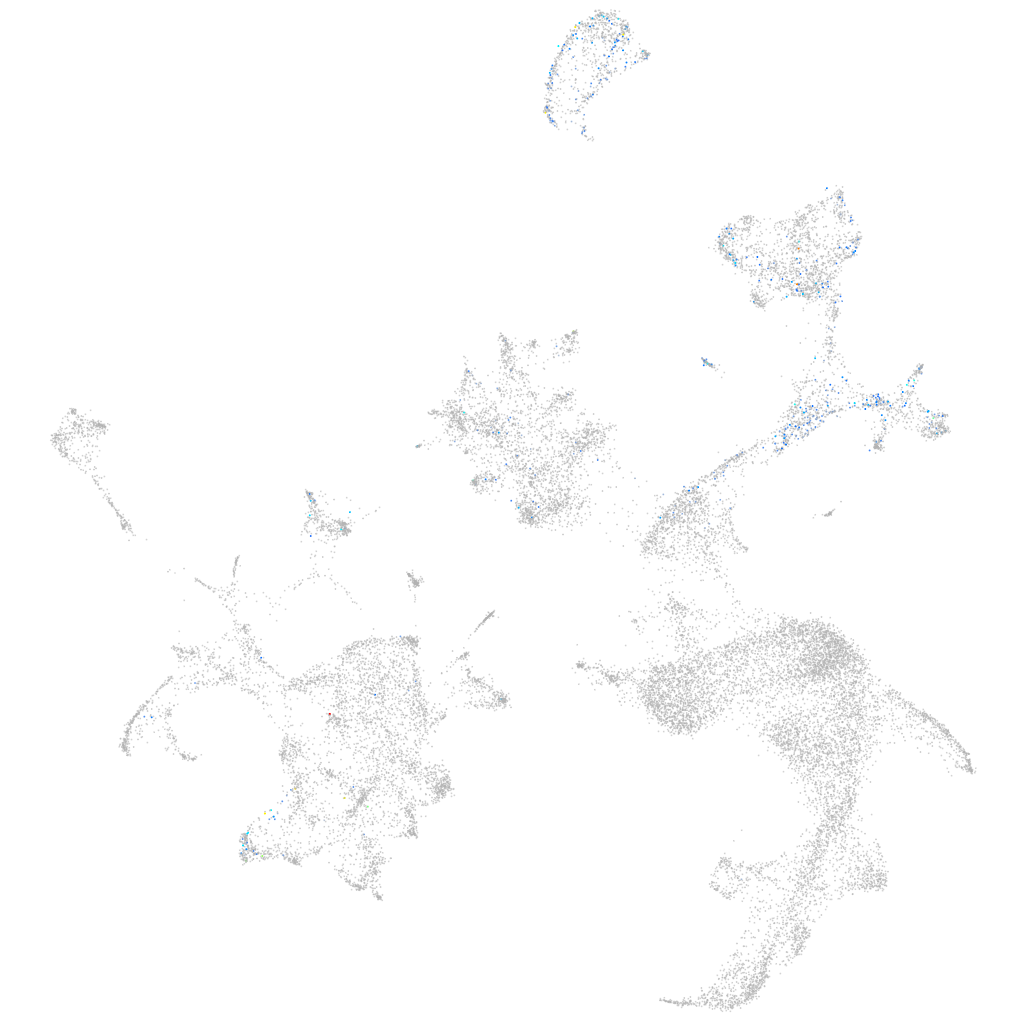
"potassium voltage-gated channel, KQT-like subfamily, member 5b"
ZFIN 
Other cell groups


all cells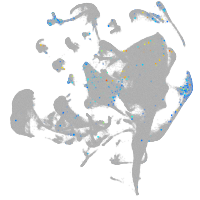 |
blastomeres |
PGCs |
endoderm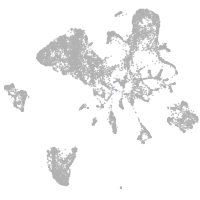 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 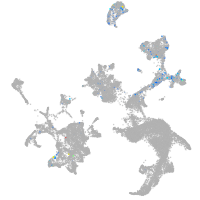 |
pronephros 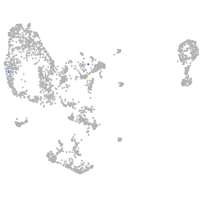 |
neural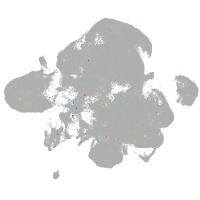 |
spinal cord / glia |
eye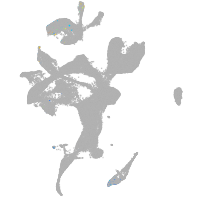 |
taste / olfactory |
otic / lateral line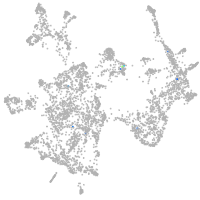 |
epidermis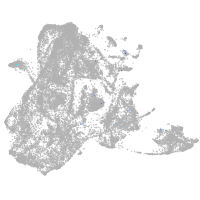 |
periderm |
fin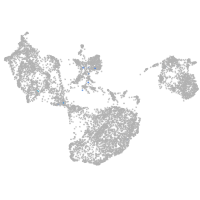 |
ionocytes / mucous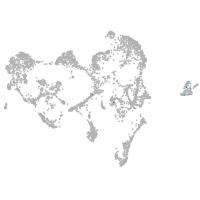 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| duox2 | 0.166 | hbae3 | -0.102 |
| laptm5 | 0.160 | hbbe2 | -0.101 |
| cebpb | 0.159 | hbbe1.3 | -0.100 |
| coro1a | 0.158 | hbae1.3 | -0.099 |
| arpc1b | 0.152 | hbbe1.1 | -0.097 |
| pfn1 | 0.152 | hbae1.1 | -0.096 |
| dusp2 | 0.147 | cahz | -0.094 |
| si:dkey-262k9.4 | 0.143 | hbbe1.2 | -0.087 |
| tnfaip8l2a | 0.143 | hemgn | -0.086 |
| cd7al | 0.142 | fth1a | -0.084 |
| arhgdig | 0.141 | alas2 | -0.082 |
| rac2 | 0.140 | blvrb | -0.080 |
| wasb | 0.139 | si:ch211-250g4.3 | -0.079 |
| ptprc | 0.138 | slc4a1a | -0.077 |
| wasa | 0.137 | nt5c2l1 | -0.077 |
| dub | 0.137 | zgc:56095 | -0.076 |
| fam49ba | 0.133 | creg1 | -0.076 |
| elf1 | 0.132 | zgc:163057 | -0.073 |
| rgs13 | 0.132 | tspo | -0.072 |
| glipr1a | 0.130 | epb41b | -0.071 |
| itgae.2 | 0.130 | si:ch211-207c6.2 | -0.068 |
| grap2b | 0.130 | nmt1b | -0.068 |
| eno3 | 0.129 | tmod4 | -0.067 |
| nrros | 0.129 | si:ch211-227m13.1 | -0.061 |
| si:cabz01036022.1 | 0.128 | plac8l1 | -0.060 |
| cotl1 | 0.127 | sptb | -0.059 |
| syk | 0.126 | hbae5 | -0.057 |
| il1b | 0.126 | hbbe3 | -0.057 |
| lta4h | 0.125 | tfr1a | -0.055 |
| si:rp71-68n21.9 | 0.125 | prdx2 | -0.055 |
| ccr9a | 0.124 | rfesd | -0.054 |
| plek | 0.124 | lmo2 | -0.053 |
| ncf1 | 0.124 | selenoj | -0.052 |
| scinlb | 0.124 | uros | -0.051 |
| si:ch73-248e21.5 | 0.123 | si:dkey-240n22.3 | -0.051 |