"potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3"
ZFIN 
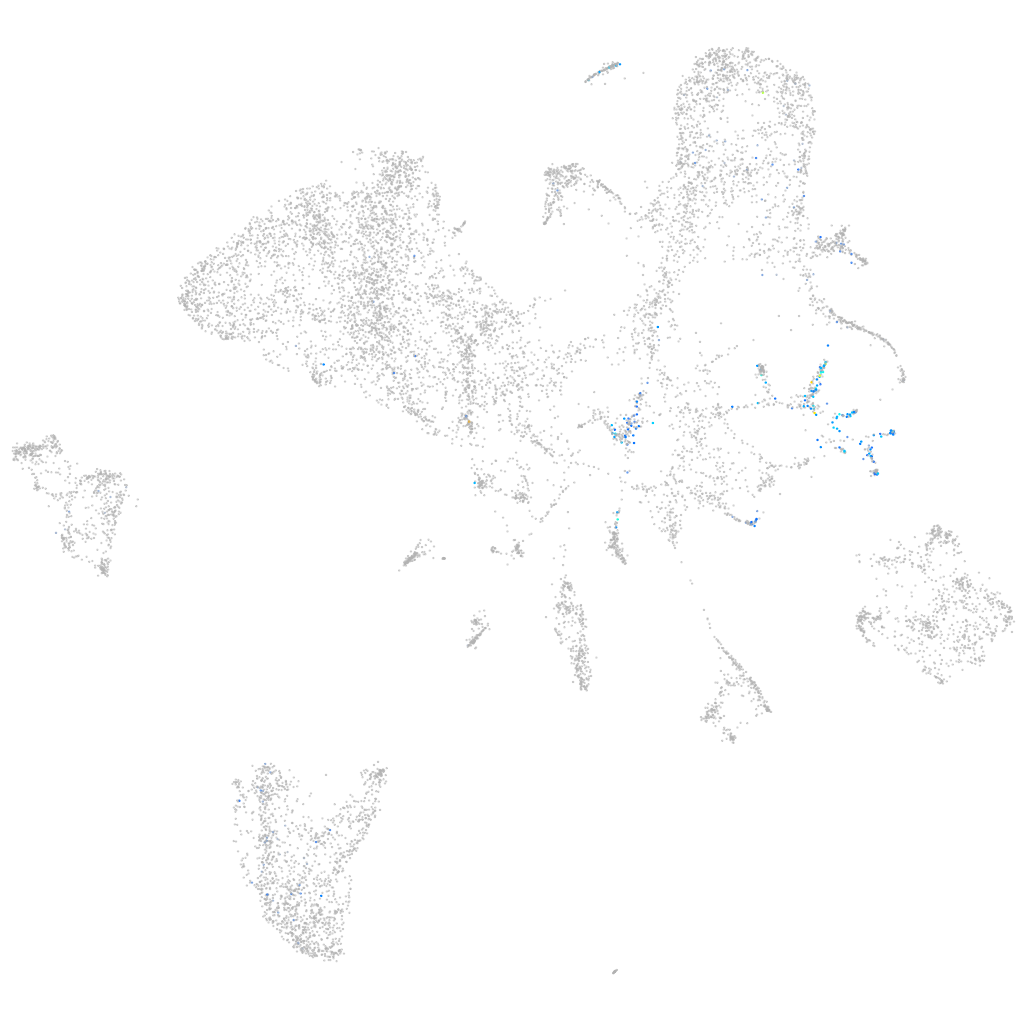
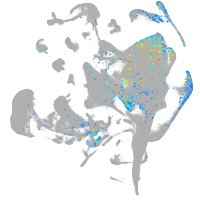
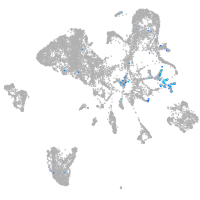
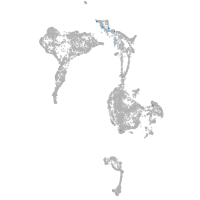
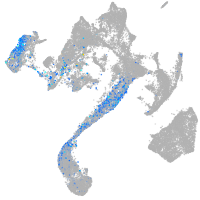
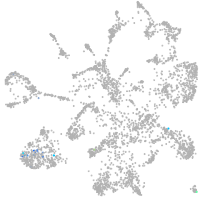
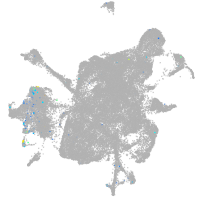
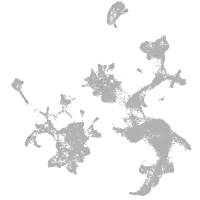
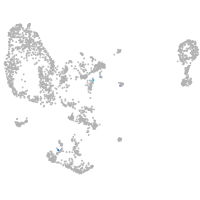
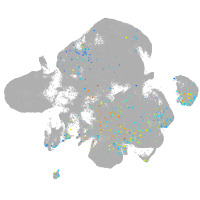
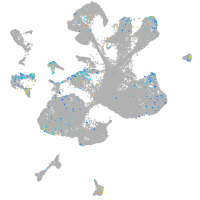
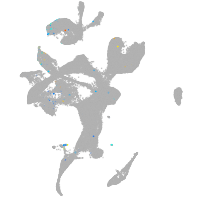
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.403 | gamt | -0.110 |
| scg3 | 0.348 | aldob | -0.106 |
| arxa | 0.337 | gapdh | -0.105 |
| isl1 | 0.329 | ahcy | -0.104 |
| neurod1 | 0.319 | bhmt | -0.099 |
| si:dkey-153k10.9 | 0.319 | fabp3 | -0.097 |
| pcsk2 | 0.317 | si:dkey-16p21.8 | -0.095 |
| kcnj11 | 0.315 | gatm | -0.095 |
| c2cd4a | 0.312 | apoa1b | -0.093 |
| abcc8 | 0.310 | fbp1b | -0.093 |
| rprmb | 0.309 | apoc1 | -0.093 |
| pcsk1nl | 0.307 | mat1a | -0.092 |
| syt1a | 0.293 | aldh6a1 | -0.092 |
| insm1a | 0.278 | hdlbpa | -0.092 |
| rims2a | 0.276 | apoc2 | -0.090 |
| nucb2b | 0.276 | glud1b | -0.090 |
| cacna1c | 0.275 | apoa4b.1 | -0.089 |
| pax6b | 0.273 | afp4 | -0.088 |
| tspan7b | 0.273 | mgst1.2 | -0.087 |
| gpc1a | 0.270 | cx32.3 | -0.087 |
| ghrl | 0.268 | gstt1a | -0.086 |
| vat1 | 0.268 | tfa | -0.086 |
| rasd4 | 0.267 | agxta | -0.085 |
| sst2 | 0.265 | apoa2 | -0.085 |
| dkk3b | 0.262 | agxtb | -0.084 |
| tango2 | 0.262 | phyhd1 | -0.083 |
| si:ch73-287m6.1 | 0.260 | wu:fj16a03 | -0.082 |
| pdyn | 0.259 | serpina1 | -0.082 |
| mir7a-1 | 0.256 | serpina1l | -0.081 |
| rapgef4 | 0.254 | rbp2b | -0.081 |
| kcnk9 | 0.248 | aldh1l1 | -0.081 |
| LOC101885494 | 0.247 | eno3 | -0.080 |
| vamp2 | 0.246 | apobb.1 | -0.080 |
| gcga | 0.245 | suclg2 | -0.080 |
| LOC571123 | 0.243 | apom | -0.079 |