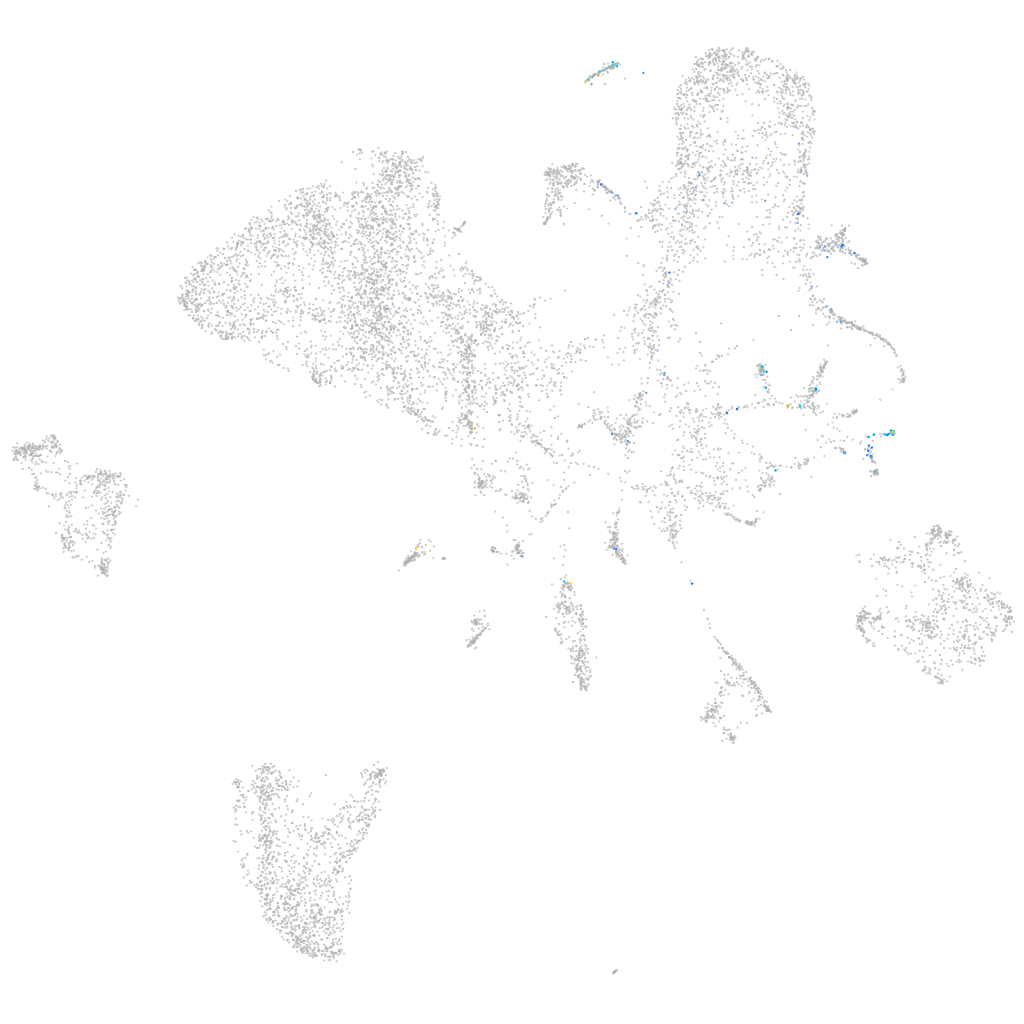
"potassium channel, subfamily K, member 3b"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 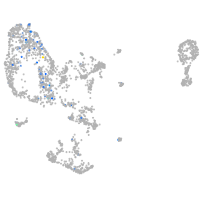 |
neural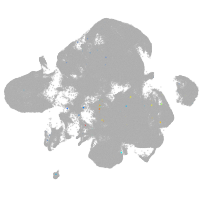 |
spinal cord / glia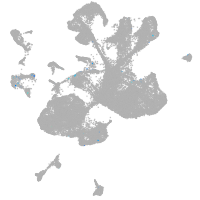 |
eye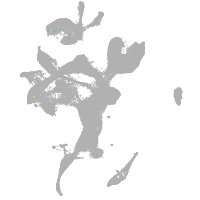 |
taste / olfactory |
otic / lateral line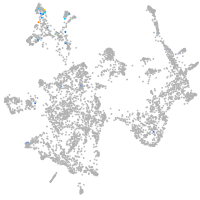 |
epidermis |
periderm |
fin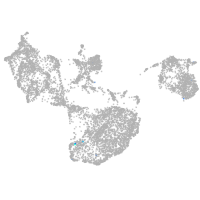 |
ionocytes / mucous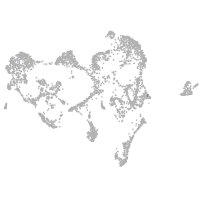 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sst2 | 0.359 | fabp3 | -0.093 |
| scg3 | 0.309 | ahcy | -0.091 |
| nmba | 0.294 | rps20 | -0.086 |
| gpr4 | 0.292 | gamt | -0.086 |
| zgc:195023 | 0.278 | gapdh | -0.081 |
| sst1.2 | 0.277 | aldob | -0.080 |
| CU019662.1 | 0.276 | aqp12 | -0.075 |
| pdyn | 0.274 | rpl15 | -0.073 |
| trpa1b | 0.271 | si:dkey-16p21.8 | -0.073 |
| gpc1a | 0.263 | gatm | -0.071 |
| neurod1 | 0.251 | bhmt | -0.070 |
| adcyap1a | 0.246 | aldh6a1 | -0.068 |
| mir7a-1 | 0.246 | mat1a | -0.066 |
| c2cd4a | 0.241 | fbp1b | -0.066 |
| kctd12.2 | 0.235 | rps3 | -0.066 |
| zgc:101731 | 0.230 | nupr1b | -0.065 |
| slc7a14a | 0.225 | hdlbpa | -0.065 |
| insm1b | 0.225 | apoa1b | -0.065 |
| nrcama | 0.223 | rps12 | -0.065 |
| gldn | 0.222 | mgst1.2 | -0.064 |
| sprn2 | 0.222 | tfa | -0.064 |
| atp8b4 | 0.220 | shmt1 | -0.063 |
| pcsk1nl | 0.219 | gstr | -0.063 |
| rgs16 | 0.215 | agxtb | -0.062 |
| kcnt1 | 0.215 | agxta | -0.062 |
| TCIM (1 of many) | 0.213 | afp4 | -0.062 |
| prlra | 0.213 | pnp4b | -0.062 |
| ACVR1C | 0.210 | aldh7a1 | -0.062 |
| hepacam2 | 0.210 | eif4ebp1 | -0.061 |
| nmu | 0.210 | apoa4b.1 | -0.061 |
| casr | 0.208 | apoc1 | -0.061 |
| disp2 | 0.208 | ttc36 | -0.061 |
| lrrc18b | 0.202 | cx32.3 | -0.061 |
| olfm2a | 0.201 | si:dkey-151g10.6 | -0.060 |
| fev | 0.201 | cpn1 | -0.060 |