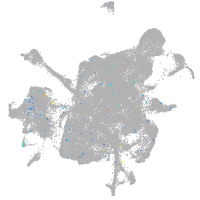
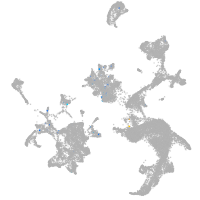
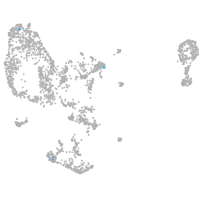
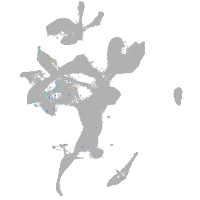
potassium inwardly rectifying channel subfamily J member 19b
ZFIN 
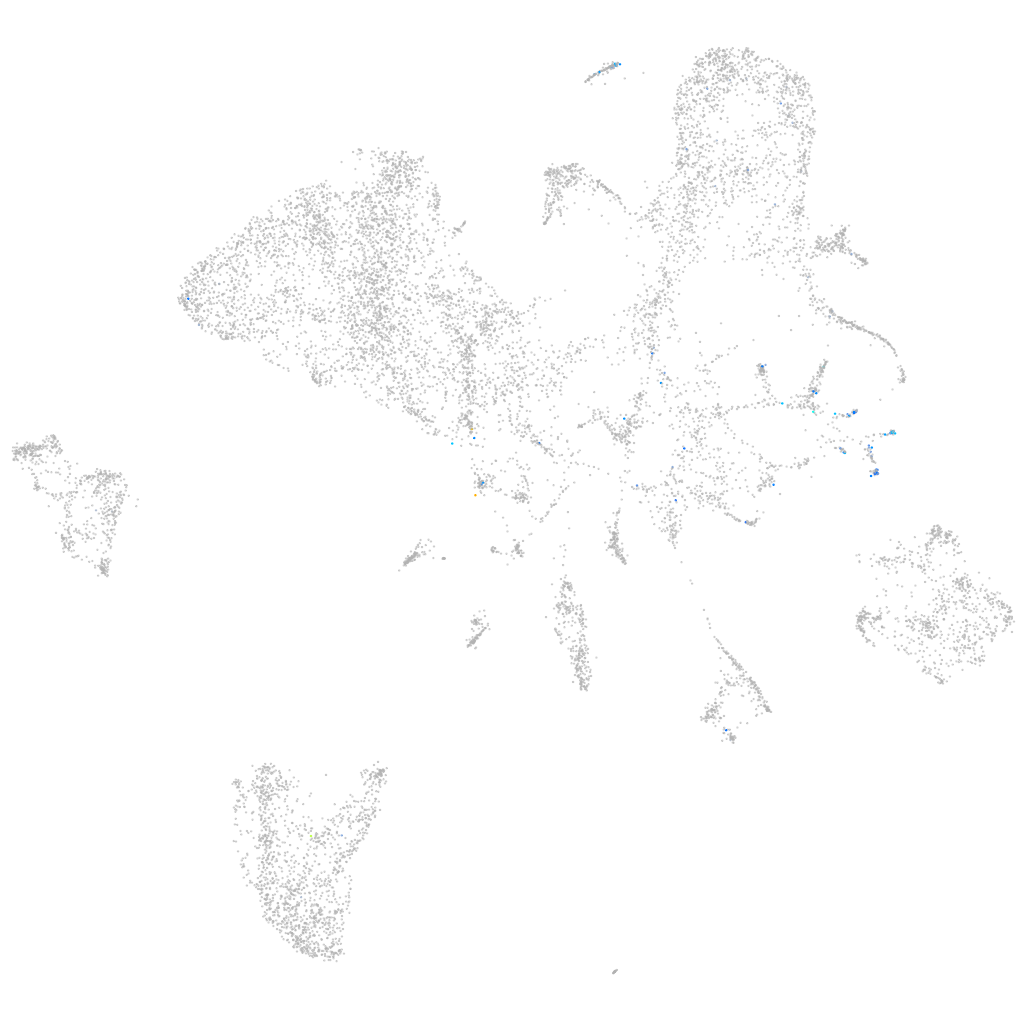
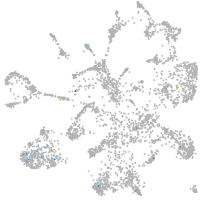
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| wu:fj39g12 | 0.308 | aldob | -0.079 |
| sprn2 | 0.263 | ahcy | -0.077 |
| si:dkeyp-74b6.2 | 0.259 | gapdh | -0.072 |
| ptchd4 | 0.258 | eno3 | -0.070 |
| GRIK2 | 0.243 | hdlbpa | -0.065 |
| scg3 | 0.224 | gamt | -0.062 |
| si:dkey-153k10.9 | 0.221 | fbp1b | -0.062 |
| pdyn | 0.219 | aldh9a1a.1 | -0.060 |
| CEP170B (1 of many) | 0.219 | gstt1a | -0.059 |
| tspan7b | 0.218 | gatm | -0.058 |
| neurod1 | 0.217 | gstp1 | -0.057 |
| pcsk1nl | 0.216 | mat1a | -0.057 |
| sst2 | 0.215 | glud1b | -0.057 |
| ins | 0.201 | gstr | -0.057 |
| gpr85 | 0.201 | gnmt | -0.057 |
| scgn | 0.198 | rplp2l | -0.056 |
| htr2aa | 0.196 | apoa4b.1 | -0.055 |
| adipoqa | 0.195 | rps20 | -0.054 |
| gabbr2 | 0.194 | adka | -0.054 |
| neurod2 | 0.194 | apoc2 | -0.053 |
| CR762493.1 | 0.194 | nupr1b | -0.053 |
| PLPP7 (1 of many) | 0.193 | bhmt | -0.053 |
| lingo1a | 0.193 | rpl3 | -0.053 |
| si:ch211-218d20.15 | 0.191 | rpl11 | -0.053 |
| gpc1a | 0.188 | agxtb | -0.053 |
| plppr4b | 0.187 | rpl15 | -0.053 |
| nmba | 0.187 | aldh7a1 | -0.053 |
| disp2 | 0.186 | apoa1b | -0.052 |
| mir7a-1 | 0.183 | apoc1 | -0.052 |
| camk1da | 0.183 | agxta | -0.051 |
| scn1lab | 0.182 | si:dkey-16p21.8 | -0.051 |
| nrsn1 | 0.182 | aqp12 | -0.051 |
| kcna2b | 0.179 | ugt1a7 | -0.051 |
| scg2a | 0.179 | acadm | -0.051 |
| impa2 | 0.179 | cx32.3 | -0.050 |