potassium inwardly rectifying channel subfamily J member 19a
ZFIN 
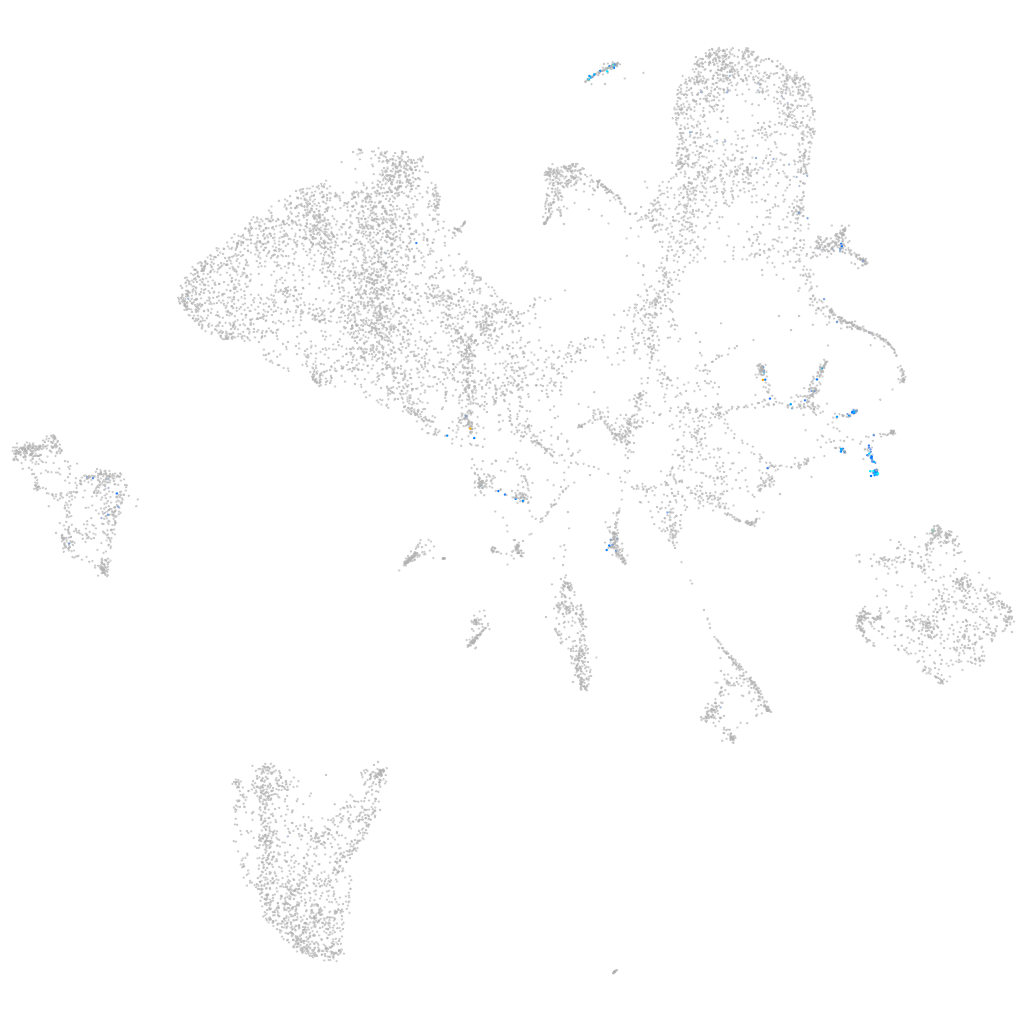
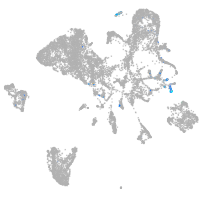
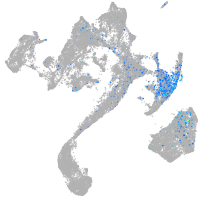
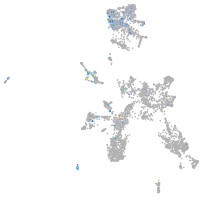
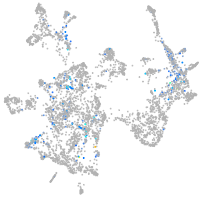
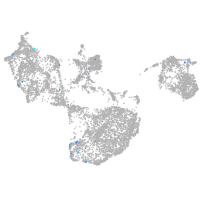
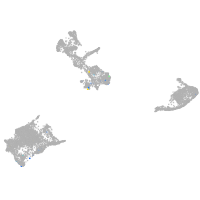
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ins | 0.406 | ahcy | -0.104 |
| ndufa4l2a | 0.398 | gapdh | -0.099 |
| ptn | 0.397 | gamt | -0.095 |
| SLC5A10 | 0.389 | aldob | -0.093 |
| slc30a2 | 0.380 | nupr1b | -0.088 |
| pcsk1 | 0.356 | hdlbpa | -0.085 |
| rims2a | 0.336 | mat1a | -0.081 |
| pcsk1nl | 0.325 | si:dkey-16p21.8 | -0.080 |
| scg3 | 0.318 | bhmt | -0.080 |
| neurod1 | 0.307 | fbp1b | -0.079 |
| cpe | 0.302 | aldh6a1 | -0.079 |
| pcsk2 | 0.298 | fabp3 | -0.079 |
| ucn3l | 0.291 | gatm | -0.077 |
| pax6b | 0.286 | rps20 | -0.077 |
| scg2a | 0.284 | gstt1a | -0.076 |
| si:dkey-153k10.9 | 0.282 | apoa1b | -0.076 |
| scgn | 0.280 | gstr | -0.075 |
| spon1b | 0.273 | cx32.3 | -0.075 |
| rgs8 | 0.272 | aldh7a1 | -0.075 |
| zgc:194990 | 0.263 | apoa4b.1 | -0.075 |
| mir7a-1 | 0.262 | hspe1 | -0.074 |
| cntnap2a | 0.252 | eno3 | -0.073 |
| gpr158a | 0.248 | agxta | -0.072 |
| g6pcb | 0.248 | shmt1 | -0.071 |
| rims2b | 0.246 | aqp12 | -0.071 |
| CABZ01118767.1 | 0.244 | agxtb | -0.070 |
| LOC100537384 | 0.243 | acadm | -0.070 |
| trpa1b | 0.241 | apoc2 | -0.069 |
| tspan7b | 0.240 | wu:fj16a03 | -0.069 |
| syt1a | 0.239 | hspd1 | -0.069 |
| scg2b | 0.236 | apoc1 | -0.068 |
| zgc:101731 | 0.235 | ppa1b | -0.068 |
| CR774179.5 | 0.233 | afp4 | -0.067 |
| disp2 | 0.233 | chchd10 | -0.067 |
| vamp2 | 0.232 | ckba | -0.066 |