potassium inwardly rectifying channel subfamily J member 11
ZFIN 
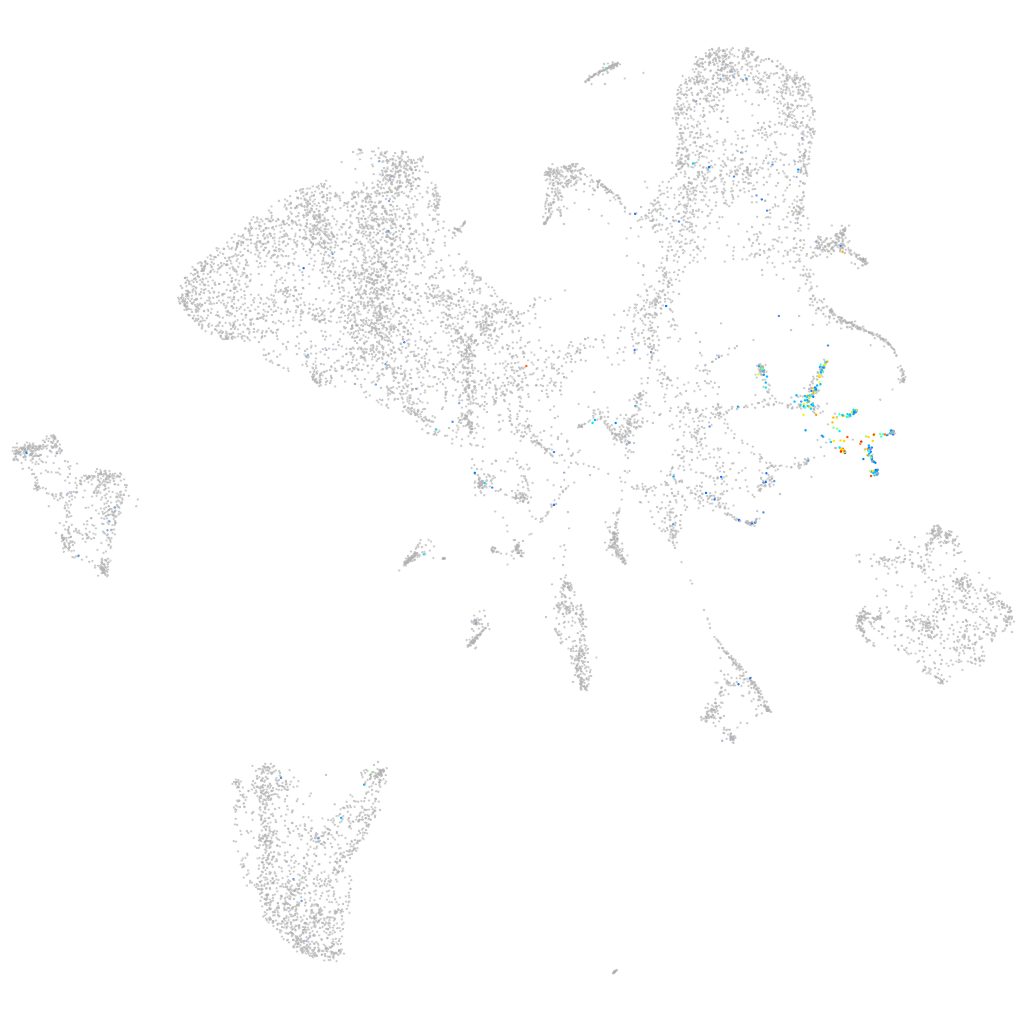
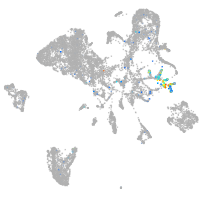
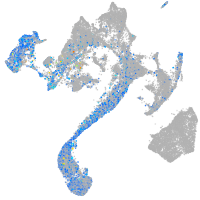
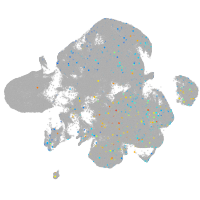
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.628 | ahcy | -0.157 |
| rprmb | 0.547 | gapdh | -0.149 |
| isl1 | 0.538 | aldob | -0.139 |
| si:dkey-153k10.9 | 0.535 | gamt | -0.133 |
| pax6b | 0.529 | gatm | -0.121 |
| scg3 | 0.524 | hdlbpa | -0.117 |
| ndufa4l2a | 0.522 | bhmt | -0.116 |
| neurod1 | 0.503 | gstp1 | -0.115 |
| pcsk2 | 0.490 | eno3 | -0.115 |
| pcsk1nl | 0.489 | aldh6a1 | -0.115 |
| SLC5A10 | 0.475 | fabp3 | -0.114 |
| syt1a | 0.464 | fbp1b | -0.112 |
| kcnk9 | 0.452 | gstr | -0.112 |
| insm1a | 0.445 | apoa4b.1 | -0.111 |
| tspan7b | 0.441 | si:dkey-16p21.8 | -0.111 |
| abhd15a | 0.437 | glud1b | -0.110 |
| mir7a-1 | 0.426 | apoa1b | -0.110 |
| arxa | 0.424 | apoc2 | -0.109 |
| fam49bb | 0.422 | mat1a | -0.109 |
| gcgb | 0.419 | rps20 | -0.109 |
| pcsk1 | 0.419 | gstt1a | -0.107 |
| ins | 0.416 | afp4 | -0.107 |
| scg2a | 0.414 | cx32.3 | -0.106 |
| vat1 | 0.409 | eif4ebp1 | -0.105 |
| si:ch73-287m6.1 | 0.408 | aldh7a1 | -0.105 |
| gpr142 | 0.406 | ppa1b | -0.104 |
| gpr22a | 0.402 | apoc1 | -0.103 |
| camk1da | 0.399 | nupr1b | -0.102 |
| rims2a | 0.399 | prdx6 | -0.101 |
| abcc8 | 0.396 | shmt1 | -0.101 |
| cacna1c | 0.394 | cebpd | -0.098 |
| ptn | 0.393 | agxta | -0.098 |
| rasd4 | 0.393 | gnmt | -0.098 |
| c2cd4a | 0.390 | apoa2 | -0.098 |
| LOC101885494 | 0.387 | mgst1.2 | -0.097 |