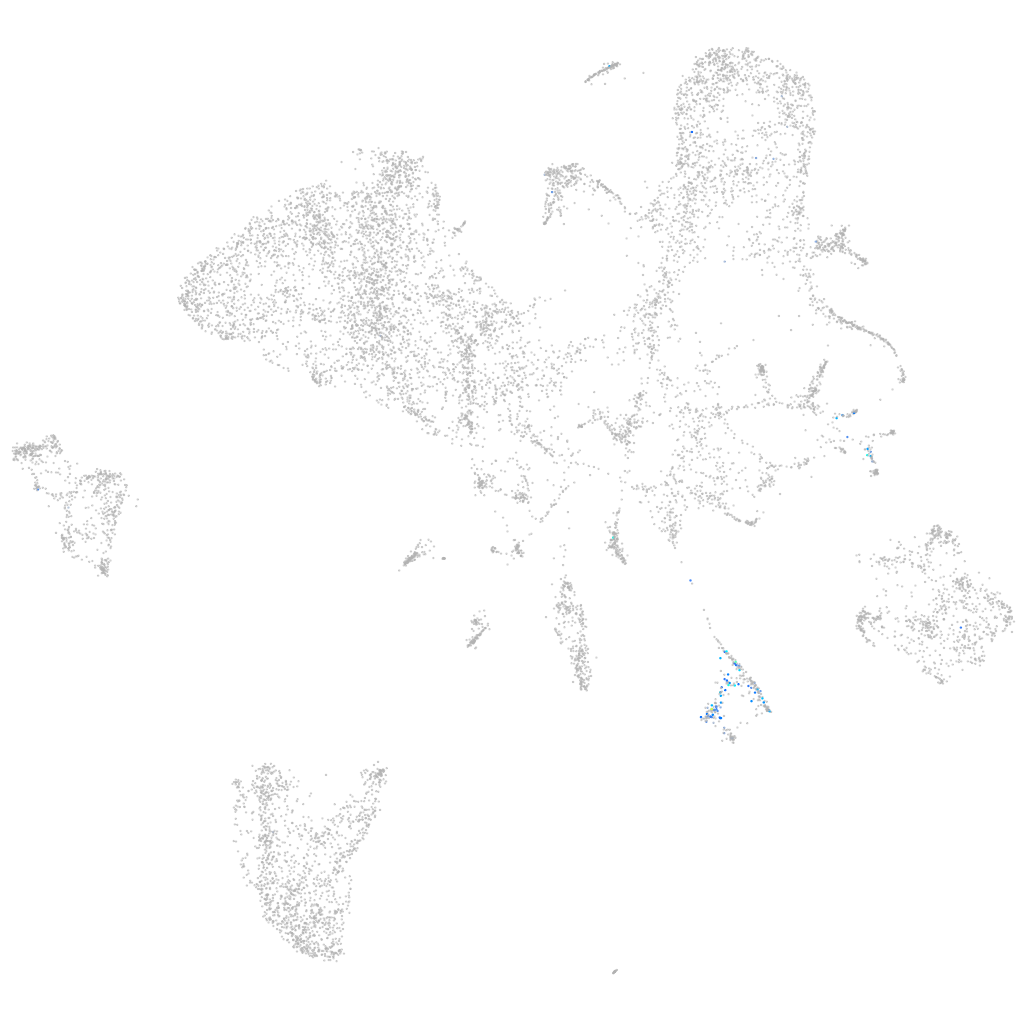
"potassium voltage-gated channel, subfamily G, member 1"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 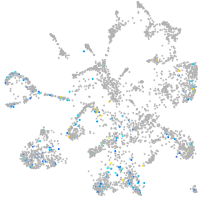 |
mesenchyme 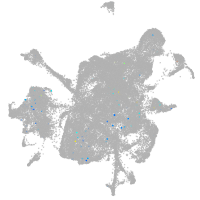 |
hematopoietic / vasculature 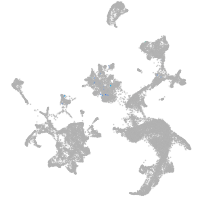 |
pronephros 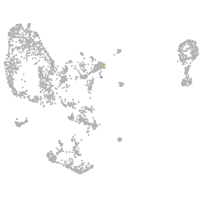 |
neural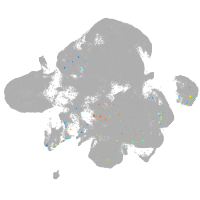 |
spinal cord / glia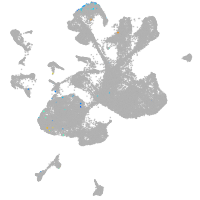 |
eye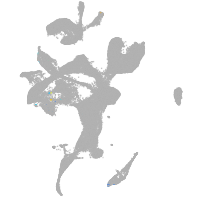 |
taste / olfactory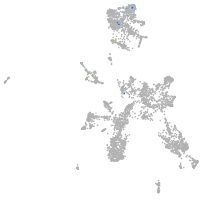 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sftpba | 0.336 | gatm | -0.072 |
| spock3 | 0.318 | gapdh | -0.072 |
| cabp2b | 0.303 | bhmt | -0.070 |
| aqp1a.1 | 0.294 | fbp1b | -0.068 |
| abca12 | 0.275 | gamt | -0.067 |
| ceacam1 | 0.275 | ahcy | -0.065 |
| sim1b | 0.274 | mat1a | -0.065 |
| sgms1 | 0.271 | apoa4b.1 | -0.065 |
| anxa3b | 0.268 | apoc2 | -0.064 |
| zgc:63831 | 0.264 | afp4 | -0.062 |
| pmp22b | 0.259 | aqp12 | -0.062 |
| mxra8b | 0.256 | apoa1b | -0.062 |
| b3gnt5a | 0.248 | gpx4a | -0.062 |
| zgc:153372 | 0.246 | si:dkey-16p21.8 | -0.058 |
| CABZ01055365.1 | 0.240 | apoc1 | -0.058 |
| rab32a | 0.235 | hspe1 | -0.057 |
| wif1 | 0.231 | aldob | -0.057 |
| LOC100330250 | 0.228 | scp2a | -0.056 |
| LOC100001846 | 0.227 | agxta | -0.056 |
| spns2 | 0.226 | apoa2 | -0.055 |
| glb1l | 0.218 | fabp3 | -0.055 |
| myl9a | 0.218 | gcshb | -0.055 |
| cygb1 | 0.216 | gstt1a | -0.055 |
| gna15.1 | 0.216 | sult2st2 | -0.055 |
| marcksl1a | 0.215 | grhprb | -0.054 |
| cd151 | 0.215 | g6pca.2 | -0.054 |
| loxa | 0.214 | agxtb | -0.054 |
| CABZ01050238.1 | 0.214 | apobb.1 | -0.054 |
| cav1 | 0.213 | dap | -0.053 |
| vwa1 | 0.213 | mdh1aa | -0.053 |
| frem2b | 0.212 | slc27a2a | -0.053 |
| col4a5 | 0.211 | lgals2b | -0.053 |
| tgfb3 | 0.211 | pnp4b | -0.052 |
| arhgap29b | 0.206 | slc16a10 | -0.052 |
| capn2a | 0.206 | ftcd | -0.052 |